Franz Baumdicker
@fbaumdicker.bsky.social
680 followers
240 following
16 posts
Mathematical and Computational Population Genetics
Microbial Evolution | Machine Learning in PopGen | CRISPR-Cas | Pangenomes | HGT
🦠 ➔ 🧬 ➔ 💻 ➔ 🤔
www.baumdickergroup.de
Posts
Media
Videos
Starter Packs
Reposted by Franz Baumdicker
Reposted by Franz Baumdicker
James Kitchens
@kitchensjn.bsky.social
· Aug 19

tskit_arg_visualizer: interactive plotting of ancestral recombination graphs
Summary: Ancestral recombination graphs (ARGs) are a complete representation of the genetic relationships between recombining lineages and are of central importance in population genetics. Recent brea...
arxiv.org
Reposted by Franz Baumdicker
Reposted by Franz Baumdicker
Fabienne Benz
@fabbenz.bsky.social
· May 15
Reposted by Franz Baumdicker
Reposted by Franz Baumdicker
Reposted by Franz Baumdicker
Anne Kupczok
@annecmg.bsky.social
· Apr 10
Reposted by Franz Baumdicker
Dan Schrider
@samurscicop.bsky.social
· Mar 24

Accessible, realistic genome simulation with selection using stdpopsim
Selection is a fundamental evolutionary force that shapes patterns of genetic variation across species. However, simulations incorporating realistic selection along heterogeneous genomes in complex de...
doi.org
Reposted by Franz Baumdicker
Franz Baumdicker
@fbaumdicker.bsky.social
· Feb 25
Reposted by Franz Baumdicker
Reposted by Franz Baumdicker
Anne Kupczok
@annecmg.bsky.social
· Feb 11
Reposted by Franz Baumdicker
Reposted by Franz Baumdicker
Franz Baumdicker
@fbaumdicker.bsky.social
· Jan 21
Reposted by Franz Baumdicker
Jakob Macke
@jakhmack.bsky.social
· Jan 20
Reposted by Franz Baumdicker
Reposted by Franz Baumdicker
Jamie Hall
@jpjhall.bsky.social
· Dec 3
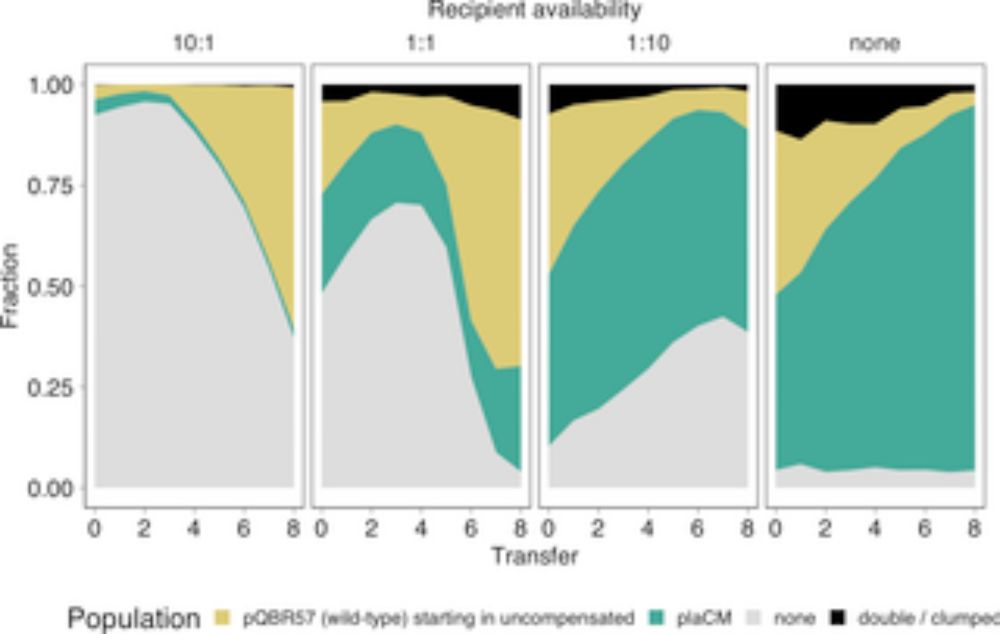
A chromosomal mutation is superior to a plasmid-encoded mutation for plasmid fitness cost compensation
Plasmids spread important traits in microbiomes but impose fitness costs on their hosts. This study of the ecological consequences of plasmid-bacterial compensatory evolution reveals benefits to bacte...
journals.plos.org