Marit Hetland
@genomarit.bsky.social
170 followers
150 following
8 posts
Bioinformatician at Stavanger University Hospital & PhD fellow at University of Bergen looking at Klebsiella genomics (and sometimes other bugs).
Posts
Media
Videos
Starter Packs
Reposted by Marit Hetland
Reposted by Marit Hetland
Reposted by Marit Hetland
Zamin Iqbal
@zaminiqbal.bsky.social
· Sep 7

Clustering of plasmid genomes for genomic epidemiology by using rearrangement distances, with pling
Integration of plasmids into genomic epidemiology is challenging, because there are no clearly defined evolving-units (equivalent to species), and because plasmids appear to evolve as much by structur...
www.biorxiv.org
Marit Hetland
@genomarit.bsky.social
· May 22

Complete genomes of 568 diverse Klebsiella pneumoniae species complex isolates from humans, animals, and marine sources in Norway from 2001 to 2020 | Microbiology Resource Announcements
Klebsiella pneumoniae species complex are opportunistic pathogens that can transmit between humans, animals,
and the environment (1). Here, we report hybrid genome assemblies of 578 (568 complete) gen...
journals.asm.org
Reposted by Marit Hetland
Reposted by Marit Hetland
Zamin Iqbal
@zaminiqbal.bsky.social
· May 19
Marit Hetland
@genomarit.bsky.social
· Apr 30
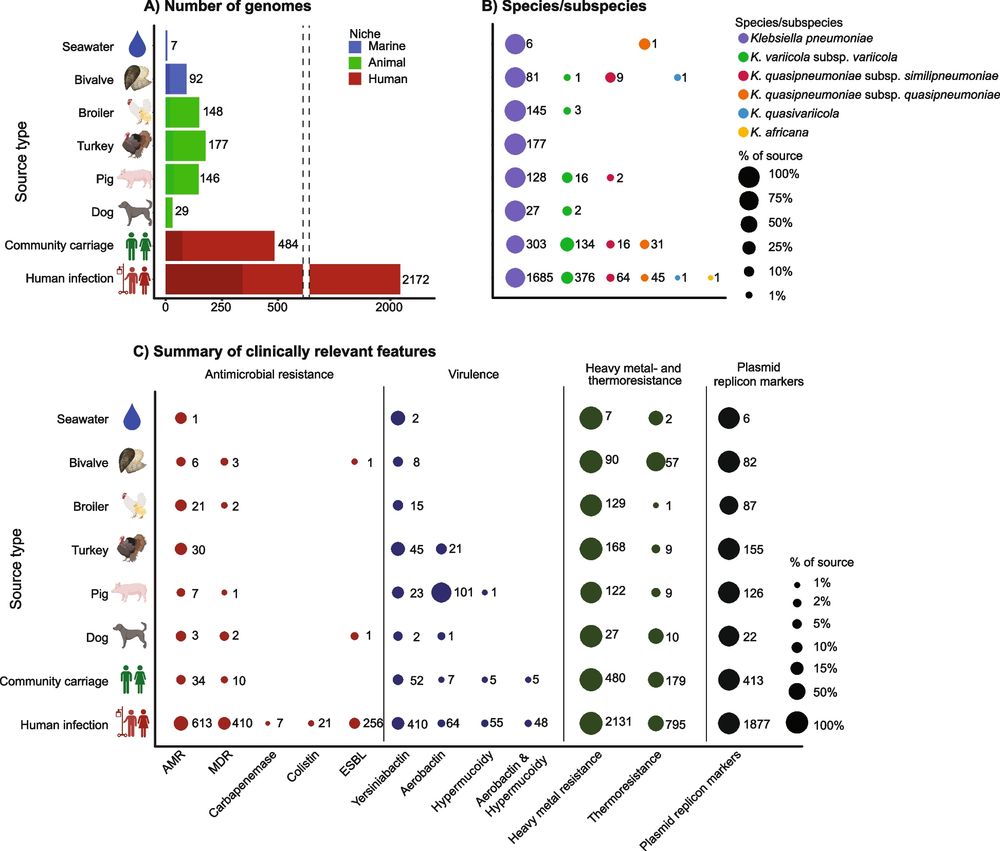
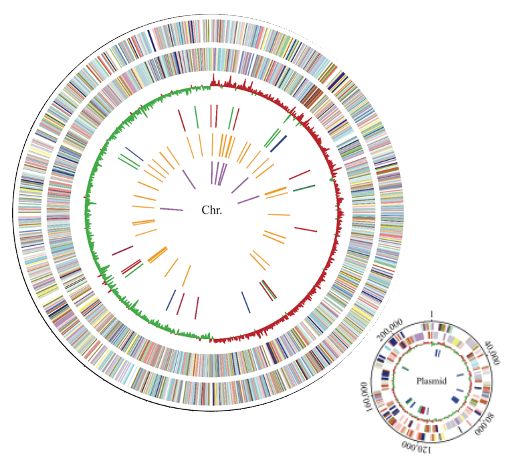
A genome-wide One Health study of Klebsiella pneumoniae in Norway reveals overlapping populations but few recent transmission events across reservoirs - Genome Medicine
Background Members of the Klebsiella pneumoniae species complex (KpSC) are opportunistic pathogens that cause severe and difficult-to-treat infections. KpSC are common in non-human niches, but the cli...
genomemedicine.biomedcentral.com
Reposted by Marit Hetland
Marit Hetland
@genomarit.bsky.social
· Nov 25
Reposted by Marit Hetland
Reposted by Marit Hetland
Willem van Schaik
@wvschaik.bsky.social
· Nov 21

A genome-wide One Health study of Klebsiella pneumoniae in Norway reveals overlapping populations but few recent transmission events across reservoirs
Members of the Klebsiella pneumoniae species complex (KpSC) are opportunistic pathogens that cause severe and difficult-to-treat infections. KpSC are common in non-human niches, but the clinical relev...
www.biorxiv.org
Marit Hetland
@genomarit.bsky.social
· Sep 13
Marit Hetland
@genomarit.bsky.social
· Sep 13
Marit Hetland
@genomarit.bsky.social
· Sep 12
Reposted by Marit Hetland
Zamin Iqbal
@zaminiqbal.bsky.social
· Oct 18