Gregor Gorjanc
@gregorgorjanc.bsky.social
390 followers
450 following
190 posts
Managing and improving populations using data science, genetics and breeding: @HighlanderLab, @RoslinInstitute, & @TheDickVet.
https://www.ed.ac.uk/roslin/highlanderlab
Posts
Media
Videos
Starter Packs
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Hanbin Lee
@epigenci.bsky.social
· Sep 4
Reposted by Gregor Gorjanc
Hannes Becher
@h-a-n.bsky.social
· Aug 28

randPedPCA: rapid approximation of principal components from large pedigrees - Genetics Selection Evolution
Background Pedigrees continue to be extremely important in agriculture and conservation genetics, with the pedigrees of modern breeding programmes easily comprising millions of records. This size can ...
doi.org
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Luis Apiolaza
@ojala.bsky.social
· Aug 26
Gregor Gorjanc
@gregorgorjanc.bsky.social
· Aug 21
Reposted by Gregor Gorjanc
Gregor Gorjanc
@gregorgorjanc.bsky.social
· Aug 11
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Reposted by Gregor Gorjanc
Jonathan Pritchard
@jkpritch.bsky.social
· Jul 28
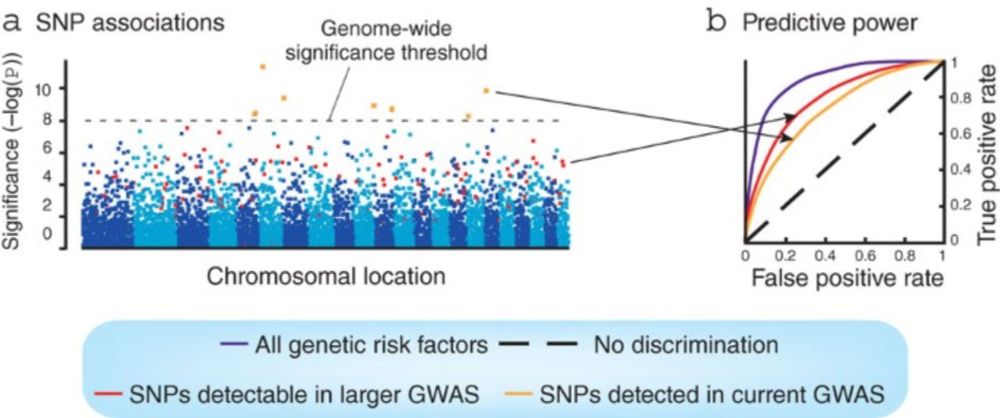
Hints of hidden heritability in GWAS - Nature Genetics
Although susceptibility loci identified through genome-wide association studies (GWAS) typically explain only a small proportion of the heritability, a classical quantitative genetic analysis now argu...
doi.org