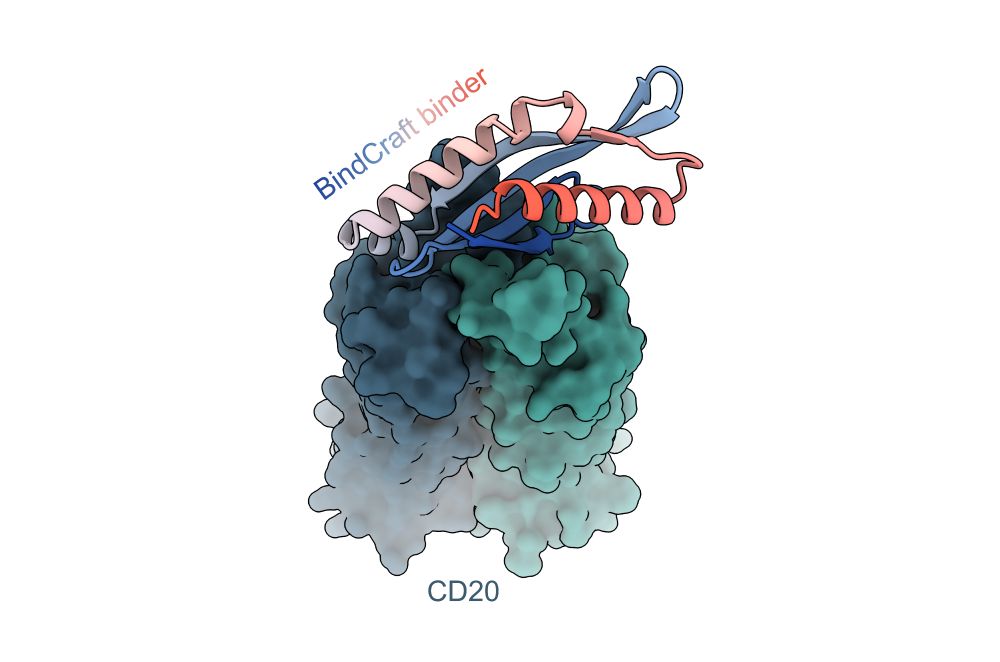
Ivan Gushchin
@ivangushchin.bsky.social
230 followers
200 following
65 posts
Structural biology and protein engineering
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Ivan Gushchin
Reposted by Ivan Gushchin
Reposted by Ivan Gushchin
BejaLab
@bejalab.bsky.social
· Sep 8

Structural basis for no retinal binding in flotillin-associated rhodopsins
Kovalev et al. determined cryo-EM structures of a flotillin-associated rhodopsin (FAR)
lacking retinal cofactor and its retinal-binding paralog, a classical light-driven
proton pump from the same bact...
www.cell.com
Reposted by Ivan Gushchin
Reposted by Ivan Gushchin
Ivan Gushchin
@ivangushchin.bsky.social
· Aug 27
Reposted by Ivan Gushchin
Ivan Gushchin
@ivangushchin.bsky.social
· Jul 26
Ivan Gushchin
@ivangushchin.bsky.social
· Jul 24
Reposted by Ivan Gushchin
Reposted by Ivan Gushchin
Reposted by Ivan Gushchin
Reposted by Ivan Gushchin
Reposted by Ivan Gushchin