Jerrin Thomas George
@jerrintgeorge.bsky.social
74 followers
140 following
13 posts
Postdoc at Wiedenheft Lab 🥼 | Former HFSP postdoctoral fellow at Sternberg Lab| 🧬 CRISPR-Cas enthusiast | Interested in mobile genetic elements
https://scholar.google.com/citations?user=sJ8ogyEAAAAJ&hl=en&oi=ao
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Jerrin Thomas George
Reposted by Jerrin Thomas George
Reposted by Jerrin Thomas George
Reposted by Jerrin Thomas George
Tugce Aktas
@aktast.bsky.social
· Jun 30
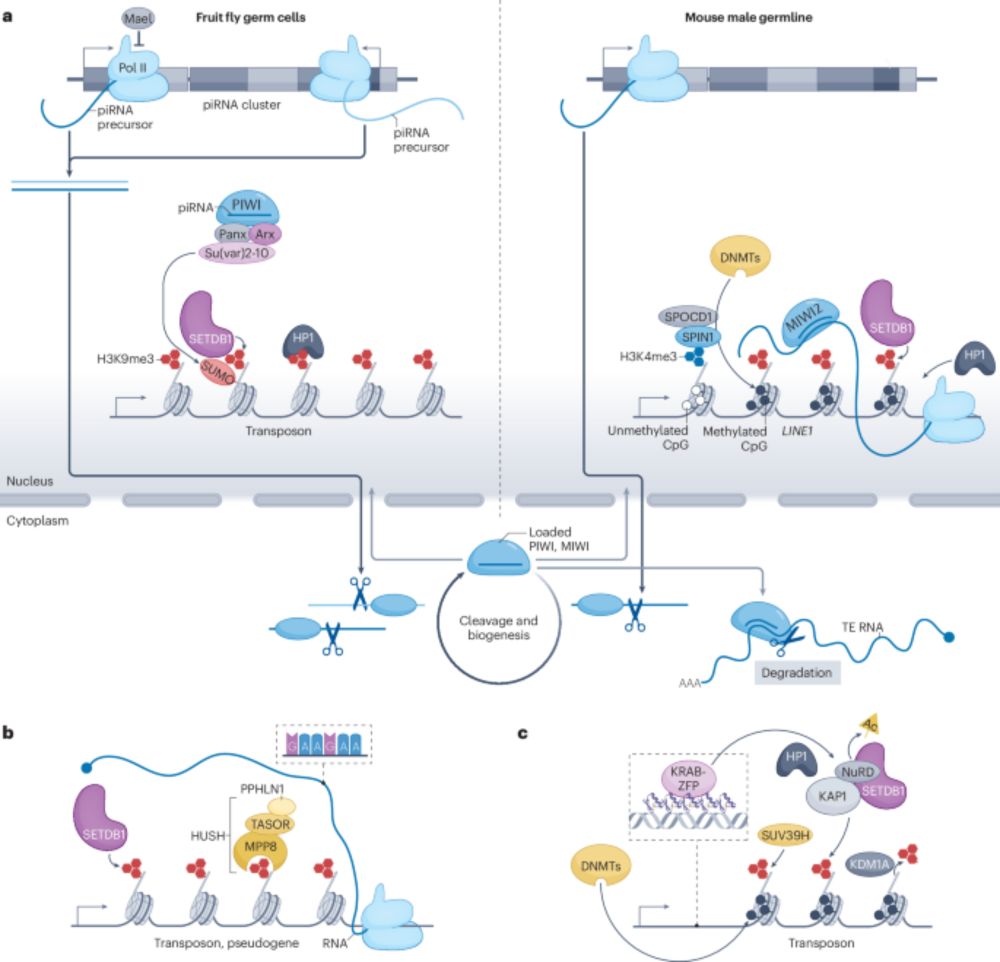
Transcriptional and post-transcriptional regulation of transposable elements and their roles in development and disease - Nature Reviews Molecular Cell Biology
Transposable elements (TEs) comprise nearly half of the human genome. This Review discusses transcriptional and post-transcriptional mechanisms that repress TE activity, how TEs escape this suppressio...
www.nature.com
Reposted by Jerrin Thomas George
Seth Shipman
@seth-shipman.bsky.social
· Jun 18
Reposted by Jerrin Thomas George
Reposted by Jerrin Thomas George
Reposted by Jerrin Thomas George
Sternberg Lab
@sternberglab.bsky.social
· May 28
Sternberg Lab
@sternberglab.bsky.social
· Mar 26

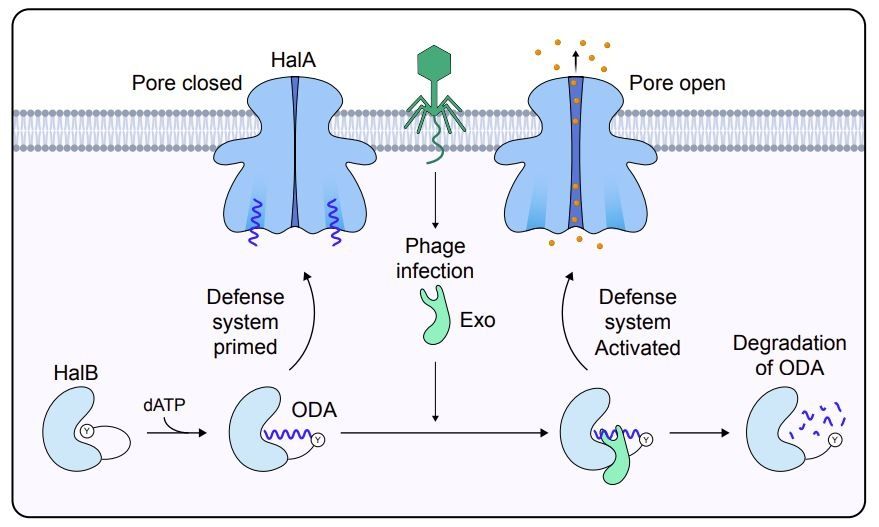
Protein-primed DNA homopolymer synthesis by an antiviral reverse transcriptase
Bacteria defend themselves from viral predation using diverse immune systems, many of which sense and target foreign DNA for degradation. Defense-associated reverse transcriptase (DRT) systems provide...
www.biorxiv.org
Reposted by Jerrin Thomas George
Reposted by Jerrin Thomas George
Reposted by Jerrin Thomas George
Liz Ortiz de Ora
@ortizdeora.bsky.social
· Apr 18
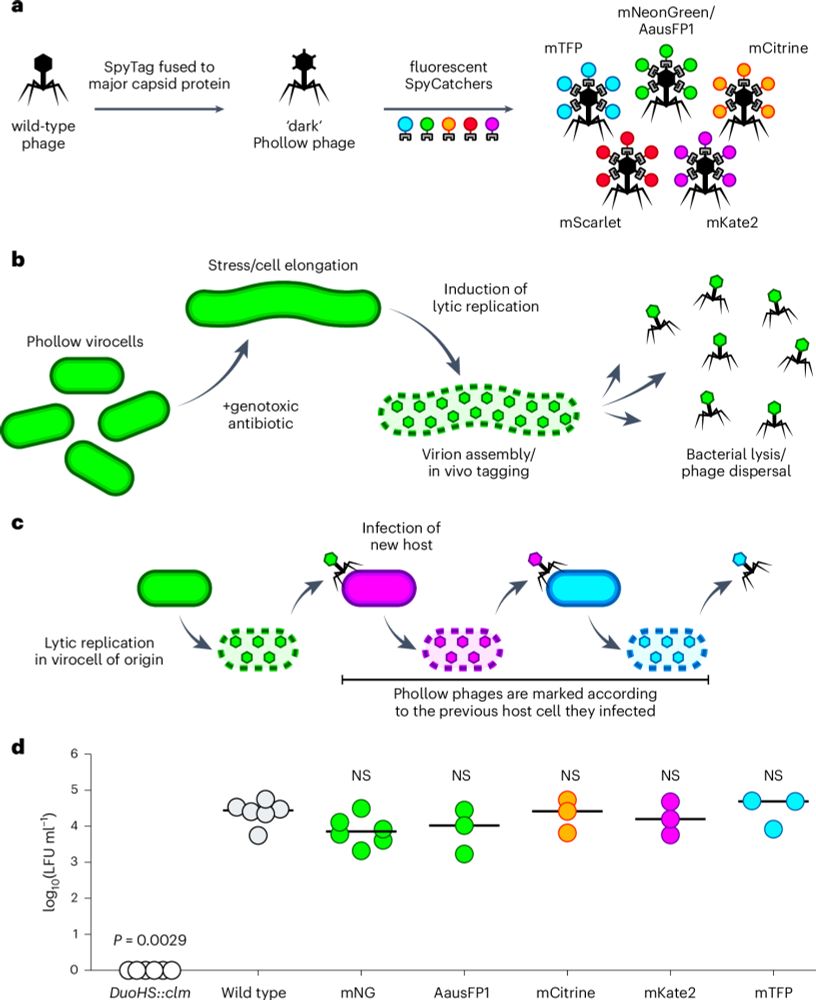
Phollow reveals in situ phage transmission dynamics in the zebrafish gut microbiome at single-virion resolution
Nature Microbiology - ‘Phollow’ is a live imaging-based fluorescence tagging approach that can track phage replication and spread in situ with single-virion resolution.
www.nature.com
Reposted by Jerrin Thomas George
Ashley Sullivan
@aesully98.bsky.social
· Mar 31

A minimal CRISPR polymerase produces decoy cyclic nucleotides to detect phage anti-defense proteins
Bacteria use antiphage systems to combat phages, their ubiquitous competitors, and evolve new defenses through repeated reshuffling of basic functional units into novel reformulations. A common theme ...
www.biorxiv.org
Reposted by Jerrin Thomas George