Jana Huisman
@jhuisman.bsky.social
930 followers
320 following
26 posts
HFSP Postdoctoral Fellow at MIT | Mobile genetic elements and microbial communities
Posts
Media
Videos
Starter Packs
Pinned
Jana Huisman
@jhuisman.bsky.social
· Aug 25

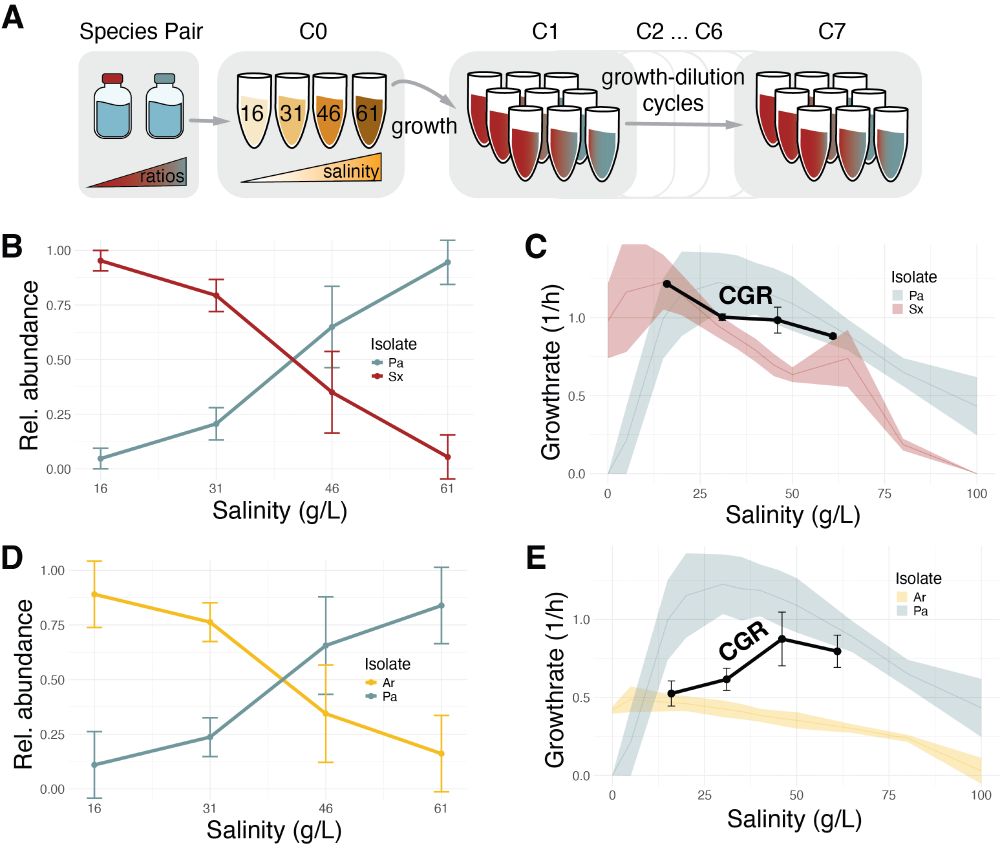
Microbial communities demonstrate robustness in stressful environments due to predictable composition shifts
Environmental stress reduces species growth rates, but its impact on the function of microbial communities is less clear. Here, we experimentally demonstrate that increasing salinity stress shifts com...
www.biorxiv.org
Reposted by Jana Huisman
Reposted by Jana Huisman
Reposted by Jana Huisman
Reposted by Jana Huisman
Reposted by Jana Huisman
Reposted by Jana Huisman
Will Smith
@willpjsmith.bsky.social
· Aug 31

Type VI secretion system activity at lethal antibiotic concentrations leads to overestimation of weapon potency
Competition assays are a mainstay of modern microbiology, offering a simple and cost-effective means to quantify microbe–microbe interactions in vitro. Here, we demonstrate a key weakness of this meth...
www.microbiologyresearch.org
Reposted by Jana Huisman
Jana Huisman
@jhuisman.bsky.social
· Aug 25
Jana Huisman
@jhuisman.bsky.social
· Aug 25

Microbial communities demonstrate robustness in stressful environments due to predictable composition shifts
Environmental stress reduces species growth rates, but its impact on the function of microbial communities is less clear. Here, we experimentally demonstrate that increasing salinity stress shifts com...
www.biorxiv.org
Jana Huisman
@jhuisman.bsky.social
· Aug 22

Transmission dynamics of Norovirus GII and Enterovirus in Switzerland during the COVID-19 pandemic (2021–2022) as evidenced in wastewater
Noroviruses and enteroviruses are major causes of endemic gastrointestinal disease associated with substantial disease burden. However, viral gastroen…
www.sciencedirect.com
Reposted by Jana Huisman
C. Brandon Ogbunu
@cbo.bsky.social
· Jul 27
Reposted by Jana Huisman
Reposted by Jana Huisman
Reposted by Jana Huisman