Kieran Campbell
@kieranrcampbell.bsky.social
3.9K followers
490 following
120 posts
https://www.camlab.ca | ML for cancer genomics + single-cell and spatial computational methods | Toronto Canada 🖥️🧬
Posts
Media
Videos
Starter Packs
Reposted by Kieran Campbell
Reposted by Kieran Campbell
Reposted by Kieran Campbell
Reposted by Kieran Campbell
Reposted by Kieran Campbell
Reposted by Kieran Campbell
Reposted by Kieran Campbell
Reposted by Kieran Campbell
Wolfgang Huber
@wkhuber.bsky.social
· Jul 29
Reposted by Kieran Campbell
Hannah Meyer
@hannahvmeyer.bsky.social
· Jul 25
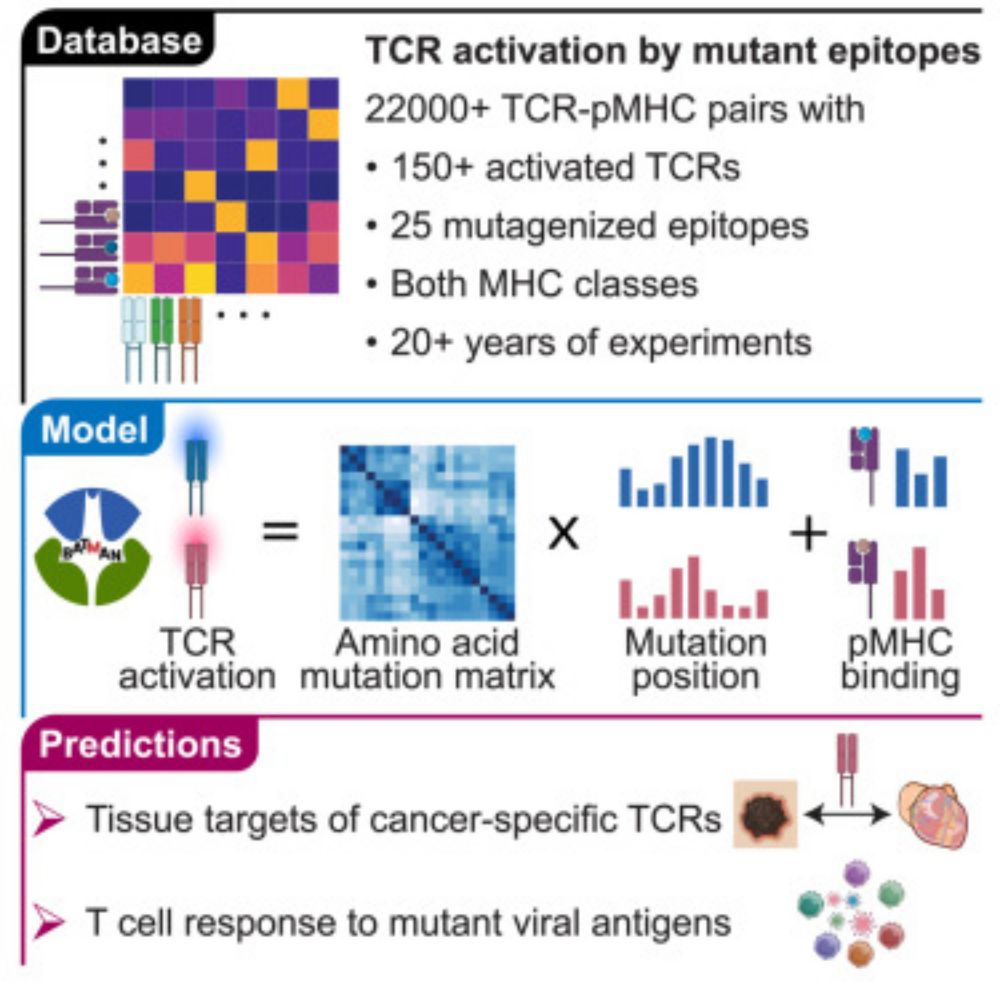
T cell receptor cross-reactivity prediction improved by a comprehensive mutational scan database
Banerjee et al. present a comprehensive T cell receptor cross-reactivity database
and a hierarchical Bayesian framework to predict T cell receptor targets. Together,
they reveal structural and biochem...
www.cell.com
Reposted by Kieran Campbell
Reposted by Kieran Campbell
Quaid Morris
@quaidmorris.bsky.social
· Jun 3

Damage and Misrepair Signatures: Compact Representations of Pan-cancer Mutational Processes
Mutational signatures of single-base substitutions (SBSs) characterize somatic mutation processes which contribute to cancer development and progression. However, current mutational signatures do not ...
www.biorxiv.org
Reposted by Kieran Campbell
Victor Javier
@vjsanchez.bsky.social
· May 27

Spatial Biology and Organoid Technologies Reveal a Potential Therapy-Resistant Cancer Stem Cell Population in Pancreatic Ductal Adenocarcinoma
Pancreatic ductal adenocarcinoma (PDAC) remains one of the deadliest malignancies with a 5-year survival rate of less than 10%. Chemotherapy is the current standard-of-care (SOC) for advanced PDAC; ho...
www.biorxiv.org