Kevin Drew
@ksdrew.bsky.social
100 followers
110 following
30 posts
Assistant Professor at University of Illinois at Chicago (UIC) in Biological Sciences focused on macromolecular assemblies.
Posts
Media
Videos
Starter Packs
Pinned
Kevin Drew
@ksdrew.bsky.social
· Jul 29

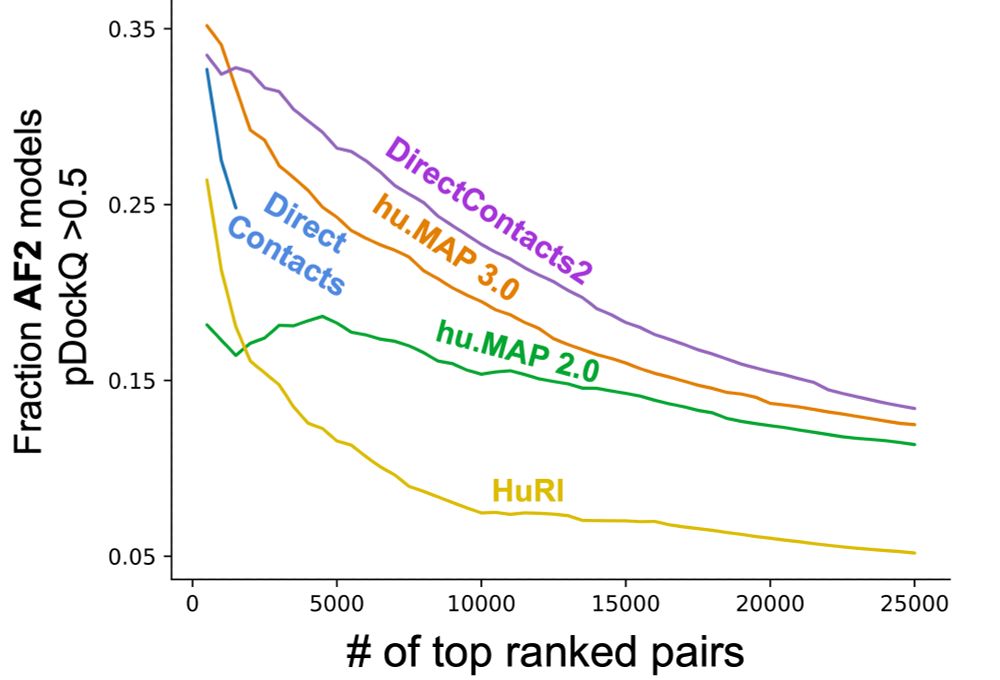
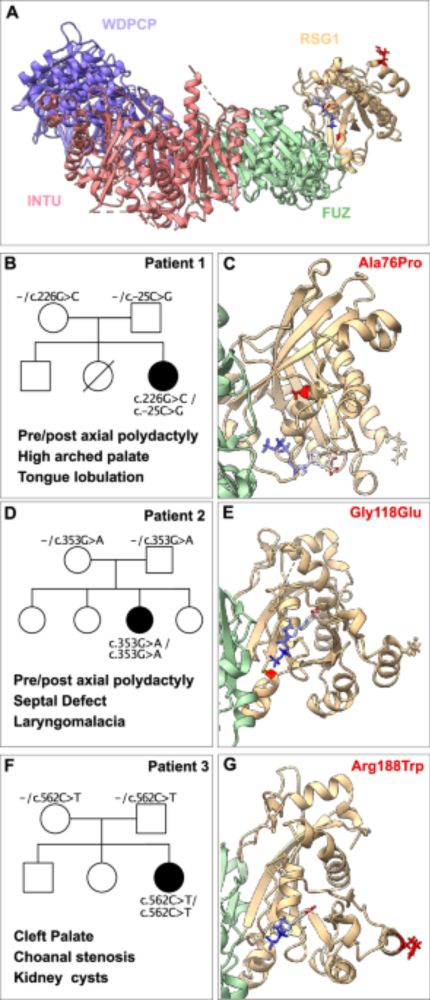
DirectContacts2: A network of direct physical protein interactions derived from high-throughput mass spectrometry experiments
Cellular function is driven by the activity proteins in stable complexes. Protein complex assembly depends on the direct physical association of component proteins. Advances in macromolecular structur...
www.biorxiv.org
Reposted by Kevin Drew
Reposted by Kevin Drew
Reposted by Kevin Drew
Reposted by Kevin Drew
Reposted by Kevin Drew
Yo Akiyama
@yoakiyama.bsky.social
· Aug 5

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
biorxiv.org
Kevin Drew
@ksdrew.bsky.social
· Jul 29
Kevin Drew
@ksdrew.bsky.social
· Jul 29
Kevin Drew
@ksdrew.bsky.social
· Jul 29
Kevin Drew
@ksdrew.bsky.social
· Jul 29
Kevin Drew
@ksdrew.bsky.social
· Jul 29
Kevin Drew
@ksdrew.bsky.social
· Jul 29

DirectContacts2: A network of direct physical protein interactions derived from high-throughput mass spectrometry experiments
Cellular function is driven by the activity proteins in stable complexes. Protein complex assembly depends on the direct physical association of component proteins. Advances in macromolecular structur...
www.biorxiv.org
Reposted by Kevin Drew
Reposted by Kevin Drew
Frank Noe
@franknoe.bsky.social
· Jul 10
Reposted by Kevin Drew