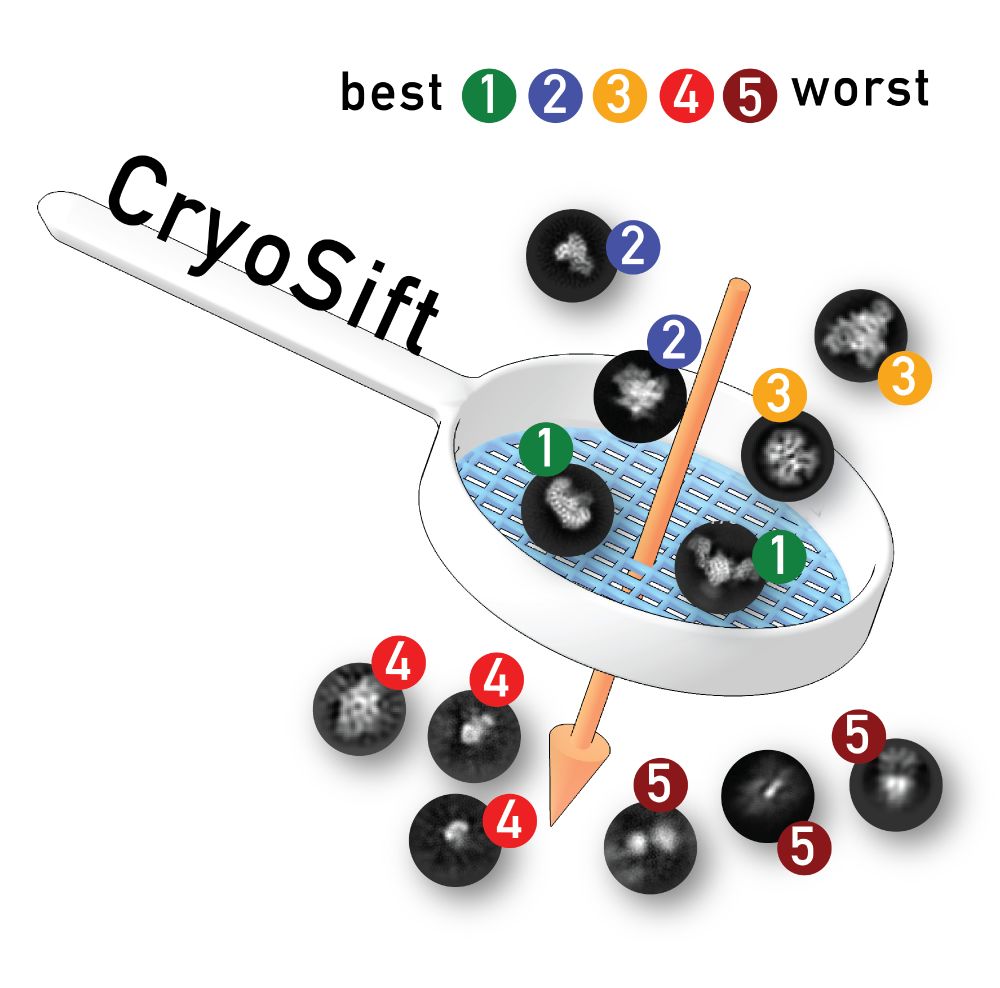
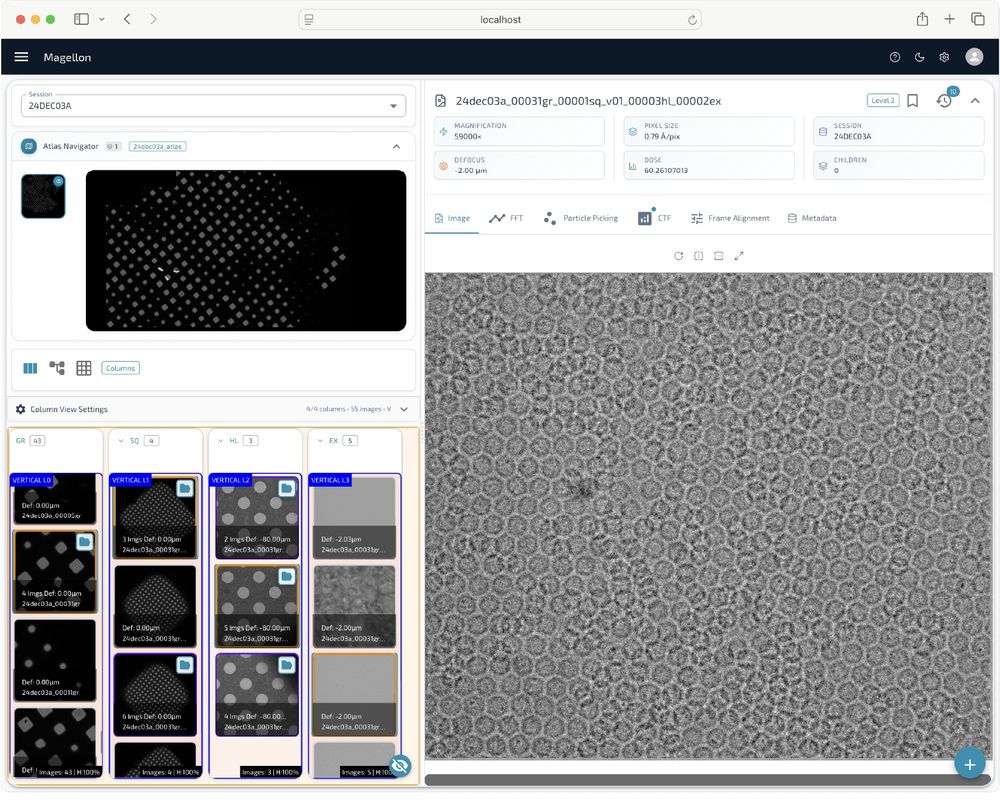
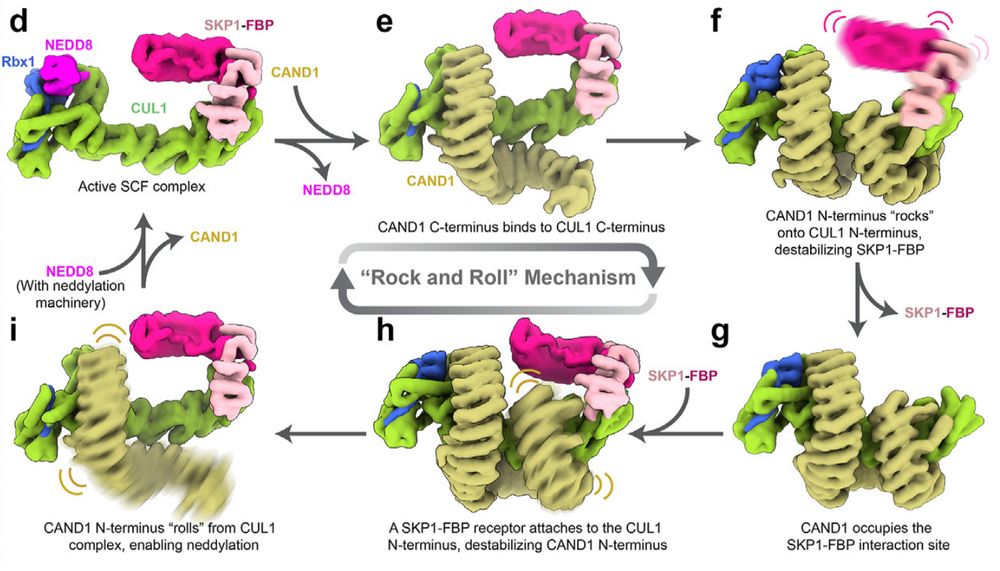
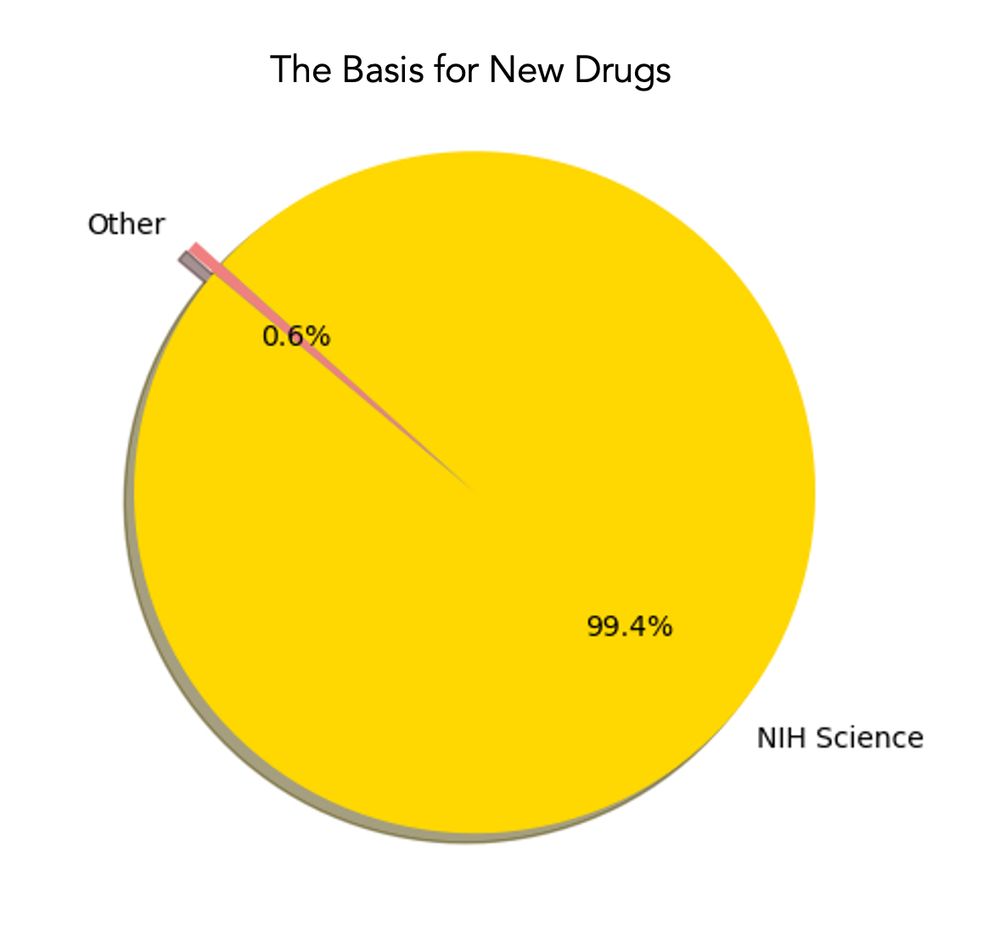
Posts
Media
Videos
Starter Packs
Reposted by Lander Lab
Reposted by Lander Lab
Reposted by Lander Lab
Reposted by Lander Lab
Reposted by Lander Lab
Lander Lab
@landerlab.bsky.social
· Apr 21
Reposted by Lander Lab
Reposted by Lander Lab
Reposted by Lander Lab
Jan-Hannes Schaefer
@jhschaef.bsky.social
· Mar 22