Posts
Media
Videos
Starter Packs
Reposted by LauraMA
Reposted by LauraMA
Reposted by LauraMA
Reposted by LauraMA
Tung Le
@tunglejic.bsky.social
· May 9

A bacterial CARD-NLR immune system controls the release of gene transfer agents
Bacteria have evolved a wide array of immune systems to detect and defend against external threats including mobile genetic elements (MGEs) such as bacteriophages, plasmids, and transposons. MGEs are ...
shorturl.at
LauraMA
@laurama-2023.bsky.social
· Apr 18
LauraMA
@laurama-2023.bsky.social
· Apr 18
LauraMA
@laurama-2023.bsky.social
· Apr 18
LauraMA
@laurama-2023.bsky.social
· Apr 18
LauraMA
@laurama-2023.bsky.social
· Apr 18
LauraMA
@laurama-2023.bsky.social
· Apr 18
LauraMA
@laurama-2023.bsky.social
· Apr 18
LauraMA
@laurama-2023.bsky.social
· Apr 18
LauraMA
@laurama-2023.bsky.social
· Apr 18
LauraMA
@laurama-2023.bsky.social
· Apr 18
Reposted by LauraMA
Reposted by LauraMA
BF Francis Ouellette
@bffo.bsky.social
· Mar 31

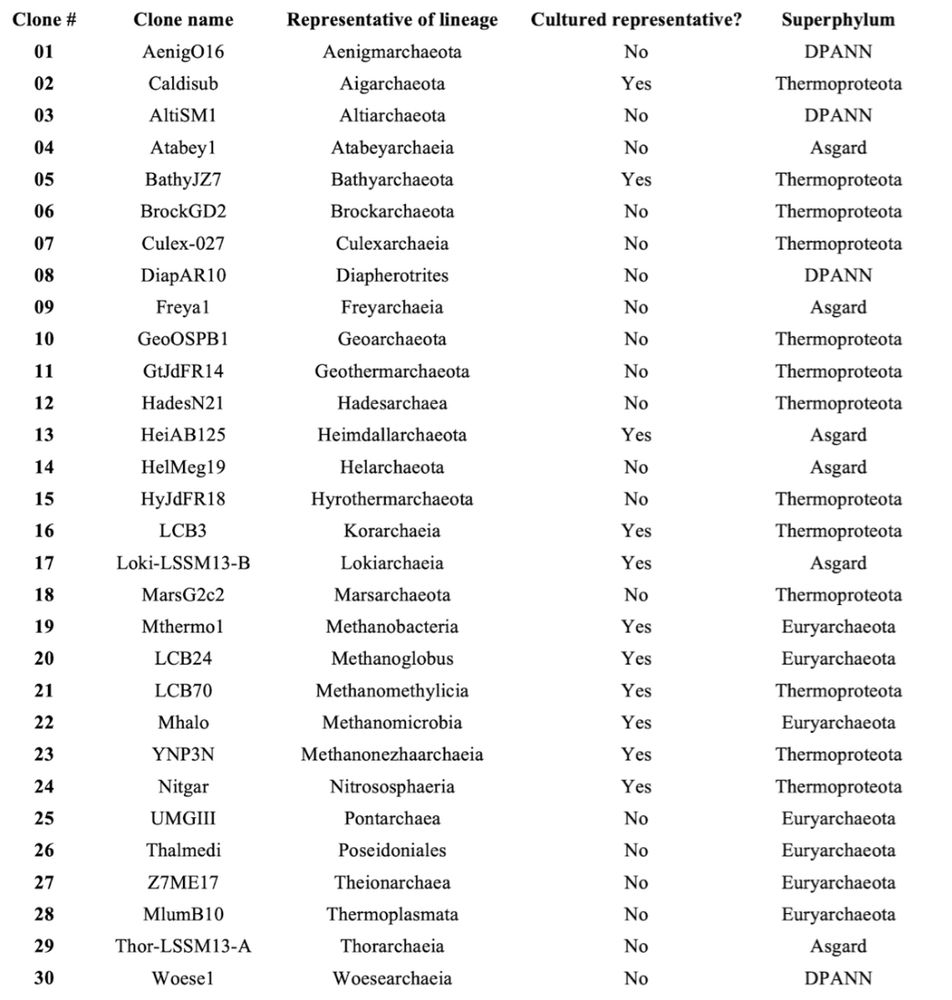
Advancing archaeal research through FAIR resource and data sharing, and inclusive community building - Communications Biology
Archaeal research and its growing importance have benefited from a community that is engaged in various collaborative efforts, which are highlighted here with examples for the sharing of resources and...
www.nature.com
Reposted by LauraMA
Reposted by LauraMA
Reposted by LauraMA
Clément Madru
@cryoclem.bsky.social
· Jan 18
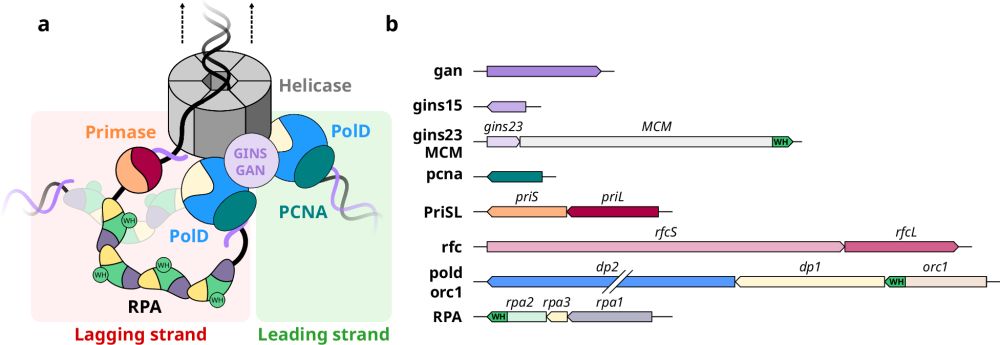
Communication between DNA polymerases and Replication Protein A within the archaeal replisome
Nature Communications - The single stranded DNA binding protein RPA plays a pivotal role in DNA replication. The archaeal RPA hosts a WH domain that interacts with the DNA primase and the...
rdcu.be