Martin
@mamueller.bsky.social
170 followers
550 following
2 posts
Postdoc, interested in translation regulation and mRNA decay
Posts
Media
Videos
Starter Packs
Reposted by Martin
Reposted by Martin
Reposted by Martin
Reposted by Martin
Reposted by Martin
Martin
@mamueller.bsky.social
· May 14

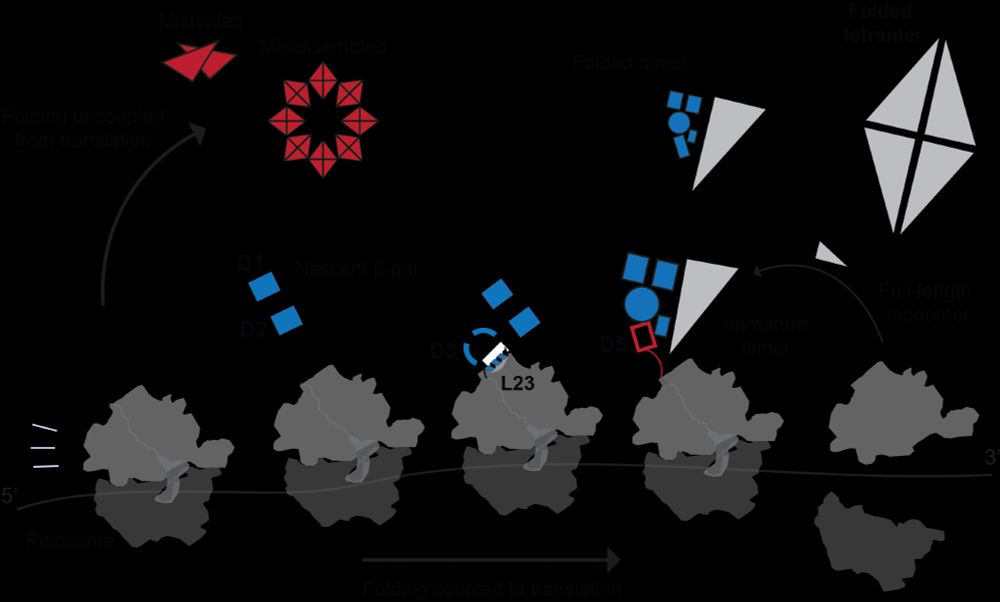
Proteome-wide determinants of co-translational chaperone binding in bacteria - Nature Communications
This study integrates ribosome profiling, single molecule methods and computational predictions to reveal that molecular chaperones bind ‘unsatisfied’ residues exposed on partial nascent folds, ration...
www.nature.com
Reposted by Martin
Yan Hu
@yanhu97.bsky.social
· Jan 23
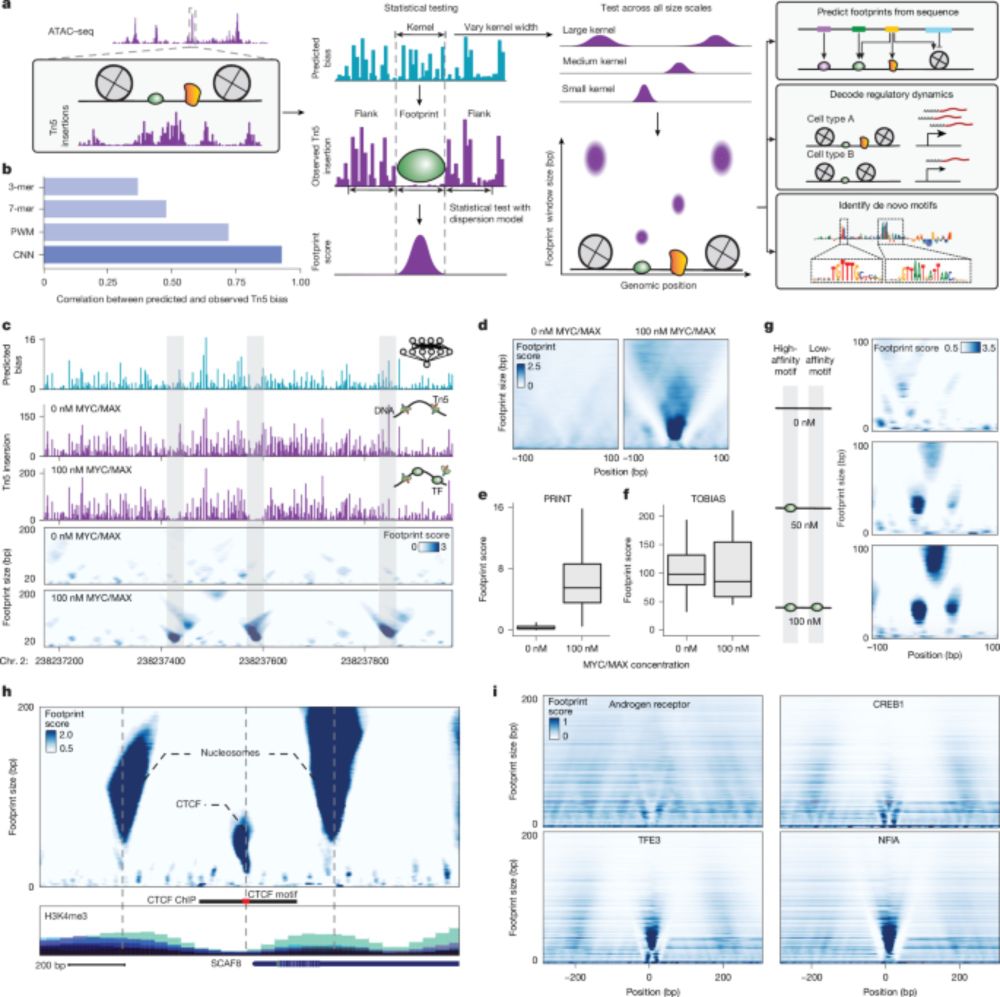
Multiscale footprints reveal the organization of cis-regulatory elements - Nature
We developed PRINT, a computational method that identifies footprints of DNA–protein interactions from bulk and single-cell chromatin accessibility data across multiple scales of protein size.
www.nature.com
Reposted by Martin
Reposted by Martin
Frances Yap
@ribo1214.bsky.social
· Dec 7

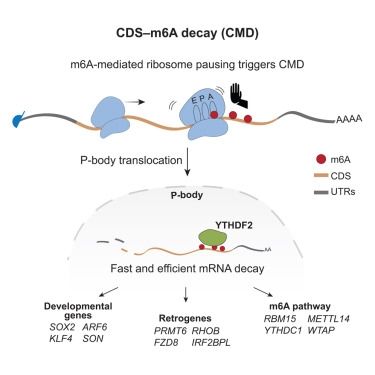
m6A sites in the coding region trigger translation-dependent mRNA decay
Zhou et al. discovered a specific role of adenosine modifications in the coding region of mRNAs. These chemical alterations slow down the movement of the ribosome during translation and trigger degradation of the corresponding mRNAs.
www.cell.com
Reposted by Martin
Reposted by Martin
Clausen Lab
@clausenlab.bsky.social
· Nov 29

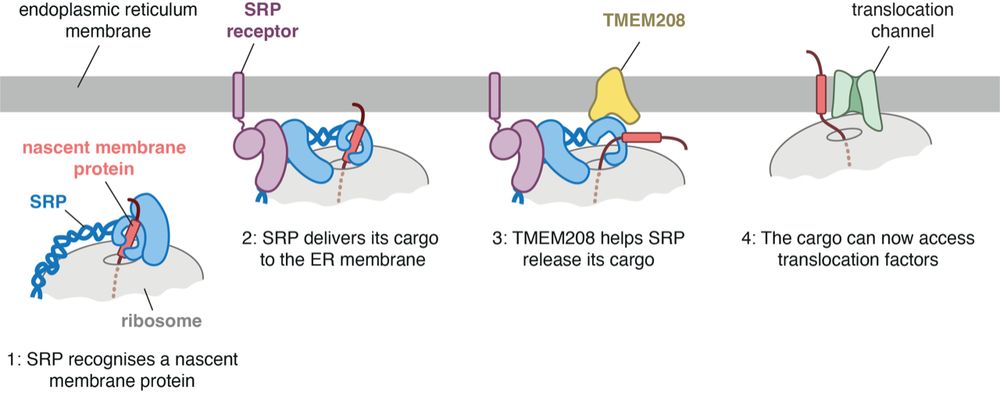
Identification of a factor that accelerates substrate release from the signal recognition particle
The eukaryotic signal recognition particle (SRP) cotranslationally recognizes the first hydrophobic segment of nascent secretory and membrane proteins and delivers them to a receptor at the endoplasmi...
www.science.org
Reposted by Martin
Reposted by Martin
Reposted by Martin