Miha Modic
@paraspeckle.bsky.social
230 followers
220 following
10 posts
Uncovering the beauty of embryonic development with functional RNA biology tools. | TT Prof at @ Carl Zeiss Center for Synthetic Genomics & KIT | http://modiclab.org/
Posts
Media
Videos
Starter Packs
Miha Modic
@paraspeckle.bsky.social
· Sep 8
Kurianlab
@kurianlab.bsky.social
· Sep 8

QKI ensures splicing fidelity during cardiogenesis by engaging the U6 tri-snRNP to activate splicing at weak 5ʹ splice sites
During organogenesis, precise pre-mRNA splicing is essential to assemble tissue architecture. Many developmentally essential exons bear weak 5'splice sites (5'SS) yet are spliced with high precision, ...
www.biorxiv.org
Reposted by Miha Modic
Trcek Lab
@trceklab.bsky.social
· Aug 30

Controlling intermolecular base pairing in Drosophila germ granules by mRNA folding and its implications in fly development - Nature Communications
Tian et al. show that RNA folding limits extensive base pairing among mRNAs in Drosophila germ granules. Instead, mRNAs interact via scattered bases exposed on the surface of folded RNAs, which may pr...
www.nature.com
Reposted by Miha Modic
Reposted by Miha Modic
Reposted by Miha Modic
Reposted by Miha Modic
Reposted by Miha Modic
Miha Modic
@paraspeckle.bsky.social
· Jul 23
Dave Shechner
@shechnerlab.bsky.social
· Jul 23
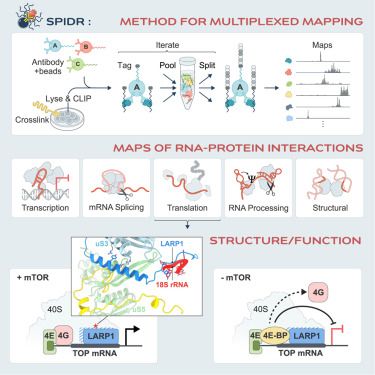
SPIDR enables multiplexed mapping of RNA-protein interactions and uncovers a mechanism for selective translational suppression upon cell stress
SPIDR, a massively multiplexed method that simultaneously maps dozens of RNA-binding
proteins to their RNA targets at single-nucleotide resolution, uncovers new RNA-protein
interactions and provides c...
www.cell.com
Miha Modic
@paraspeckle.bsky.social
· Jun 2
Miha Modic
@paraspeckle.bsky.social
· May 24
Kurianlab
@kurianlab.bsky.social
· May 24
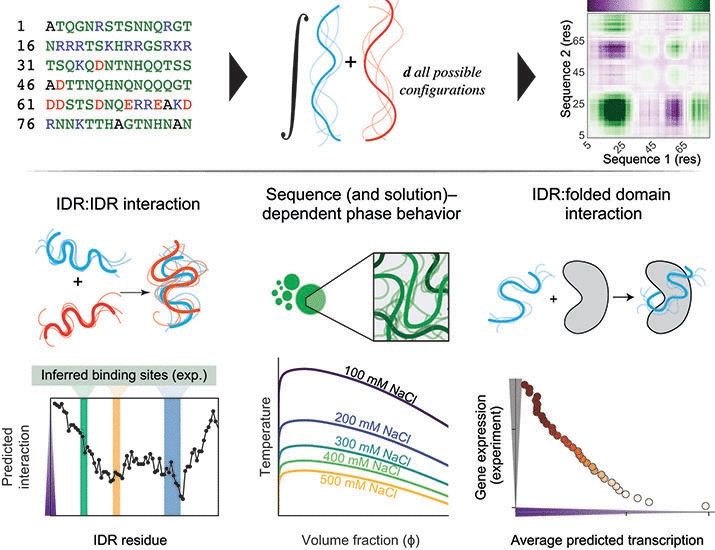
Sequence-based prediction of intermolecular interactions driven by disordered regions
Intrinsically disordered regions (IDRs) in proteins play essential roles in cellular function. A growing body of work has shown that IDRs often interact with partners in a manner that does not depend ...
www.science.org
Reposted by Miha Modic
Miha Modic
@paraspeckle.bsky.social
· Apr 18
Reposted by Miha Modic
Reposted by Miha Modic
Reposted by Miha Modic
Miha Modic
@paraspeckle.bsky.social
· Mar 6
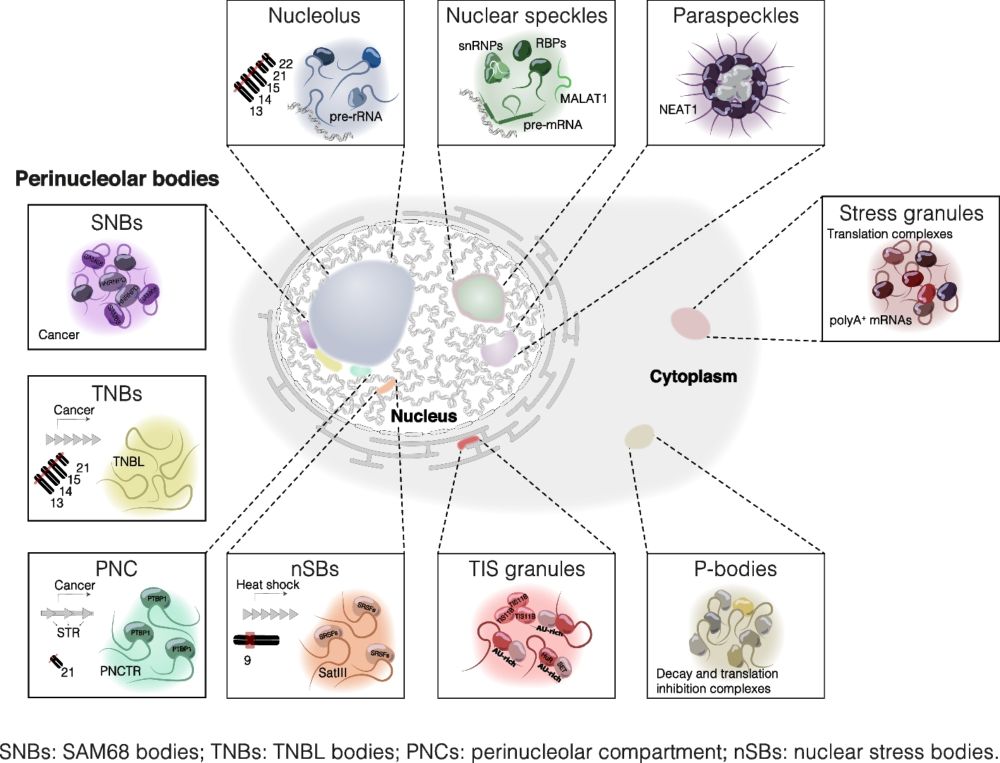
Decoding subcellular RNA localization one molecule at a time - Genome Biology
Eukaryotic cells are highly structured and composed of multiple membrane-bound and membraneless organelles. Subcellular RNA localization is a critical regulator of RNA function, influencing various bi...
genomebiology.biomedcentral.com
Reposted by Miha Modic
Reposted by Miha Modic
Miha Modic
@paraspeckle.bsky.social
· Feb 6
Miha Modic
@paraspeckle.bsky.social
· Feb 6

Integrative profiling of condensation-prone RNAs during early development
Complex RNA–protein networks play a pivotal role in the formation of many types of biomolecular condensates. How intrinsic RNA features contribute to condensate formation however remains unclear. Here...
www.biorxiv.org