Sendoel Lab
@sendoellab.bsky.social
330 followers
320 following
43 posts
Sendoel lab at the University of Zurich, alumnus of the The Rockefeller University, NYC.
We are excited about mRNA translation, stem cells and cancer.
www.sendoellab.org
Posts
Media
Videos
Starter Packs
Reposted by Sendoel Lab
Reposted by Sendoel Lab
Reposted by Sendoel Lab
Reposted by Sendoel Lab
Sendoel Lab
@sendoellab.bsky.social
· Aug 6
Reposted by Sendoel Lab
Reposted by Sendoel Lab
Reposted by Sendoel Lab
David Taborsky
@dadita.bsky.social
· Jul 25
Reposted by Sendoel Lab
Reposted by Sendoel Lab
Barbara Marte
@barbmarte.bsky.social
· May 28
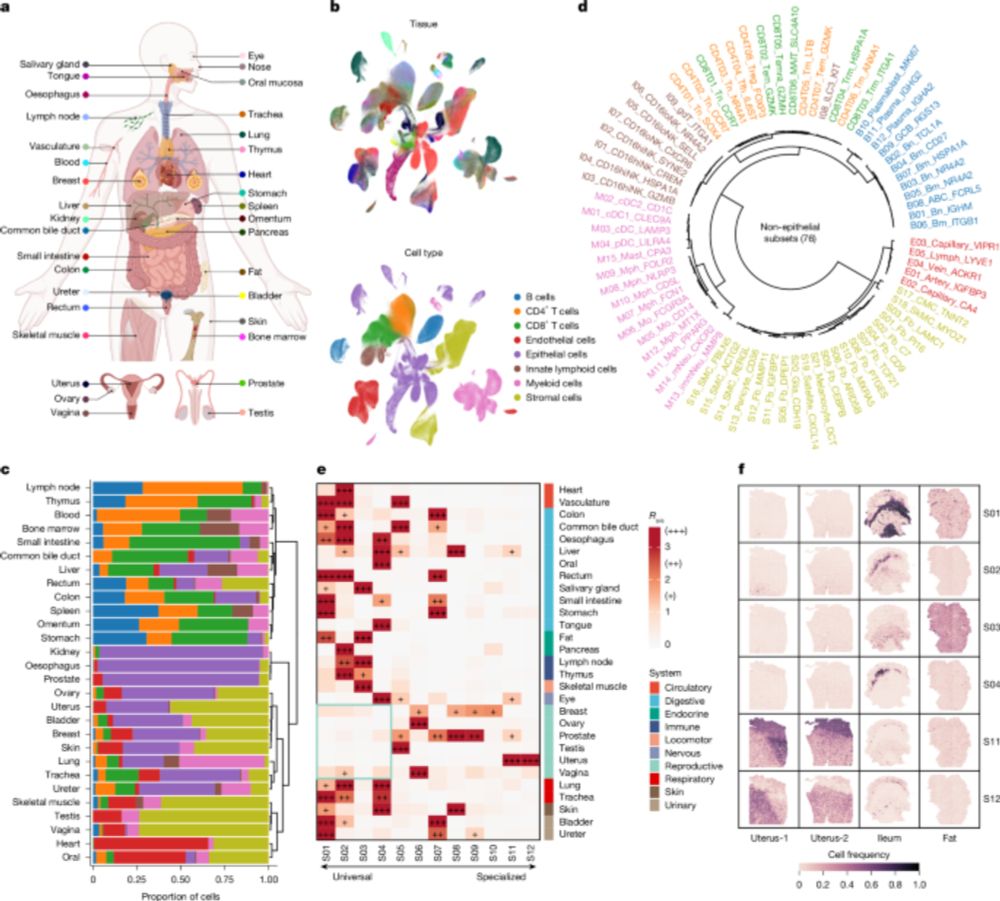
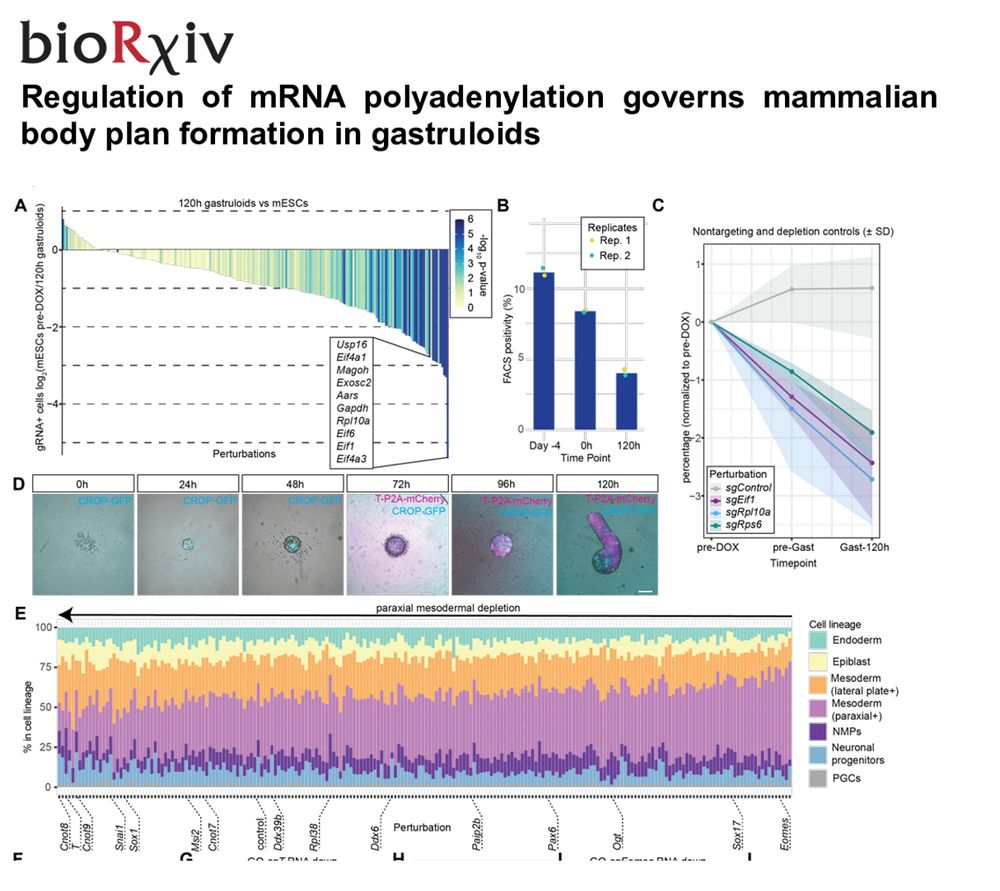
Cross-tissue multicellular coordination and its rewiring in cancer - Nature
Comprehensive single-cell transcriptomic analysis of 35 human tissues reveals 12 cross-tissue coordinated cellular modules that exhibit intramodule communication and dynamic changes with ageing, and w...
www.nature.com
Reposted by Sendoel Lab