Shiori Iida
@shiori-iida.bsky.social
100 followers
130 following
8 posts
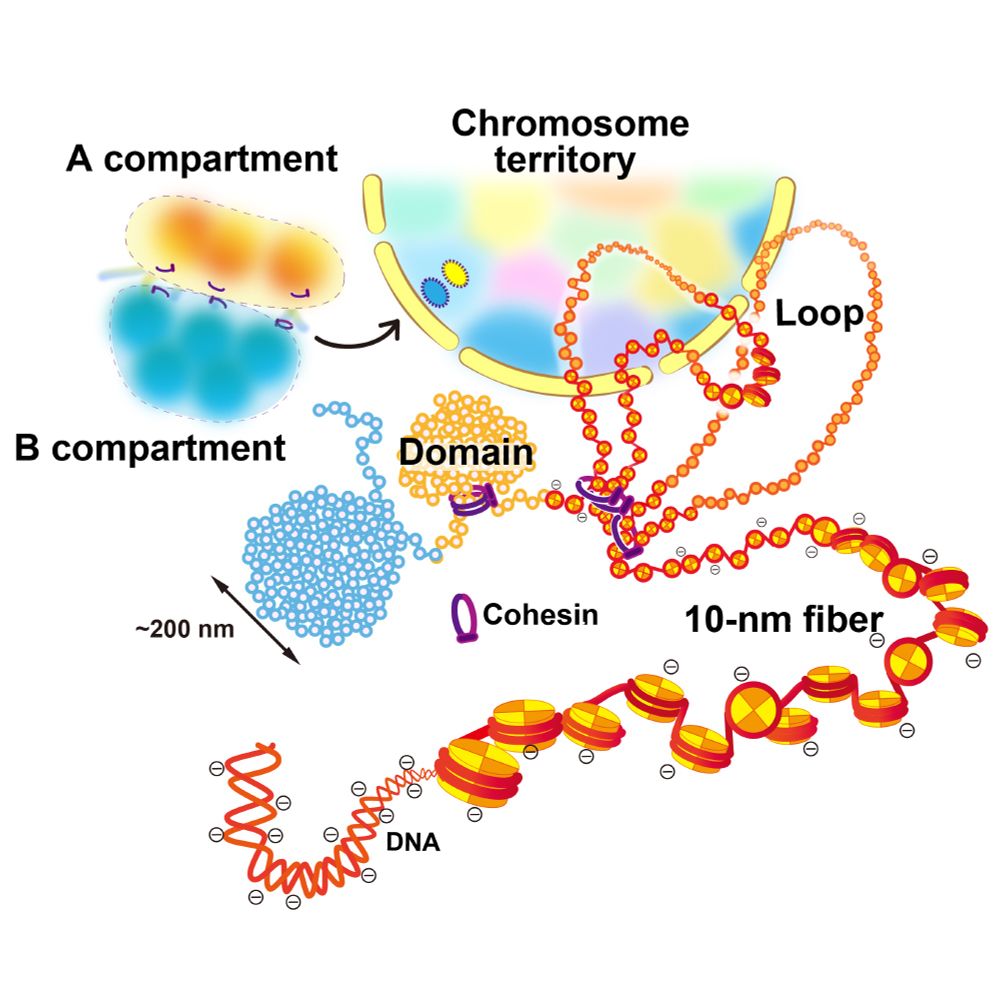
Postdoc/ PhD @kazu-maeshima.bsky.social at National Institute of Genetics, Japan / Chromatin, live-cell imaging, microscopy, recently interested in mechanobiology and cell differentiation
https://linktr.ee/shiori_iida
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Shiori Iida
Reposted by Shiori Iida
Shiori Iida
@shiori-iida.bsky.social
· Aug 30
Shiori Iida
@shiori-iida.bsky.social
· Aug 29
Shiori Iida
@shiori-iida.bsky.social
· Aug 28
Reposted by Shiori Iida
Simone Reber
@simonereber.bsky.social
· Aug 15

Conserved nucleocytoplasmic density homeostasis drives cellular organization across eukaryotes
Nature Communications - Cells can regulate their mass density. Here, the authors demonstrate how eukaryotes establish and maintain a lower density in the nucleus than in the cytoplasm via pressure...
rdcu.be
Reposted by Shiori Iida
Reposted by Shiori Iida
Reposted by Shiori Iida
Reposted by Shiori Iida
Sara Wickstrom
@sarawickstrom.bsky.social
· Apr 23
Júlia Bonjoch
@juliabonjoch.bsky.social
· Apr 23

BMAL1 and YAP cooperate to hijack enhancers and promote inflammation in the aged epidermis
Ageing is characterised by persistent low-grade inflammation that is linked to impaired tissue homeostasis and functionality. However, the molecular mechanisms driving age-associated inflammation rema...
biorxiv.org
Reposted by Shiori Iida
Reposted by Shiori Iida
Reposted by Shiori Iida
Reposted by Shiori Iida
Reposted by Shiori Iida
Reposted by Shiori Iida
Reposted by Shiori Iida