Krishna Shrinivas
@shrinivaslab.bsky.social
1.9K followers
170 following
35 posts
Asst. prof at Northwestern ChBE
Interested in how molecules and processes are organized and regulated in living cells | physics, math, engineering, and computation (mostly) for biology
shrinivaslab.com
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Krishna Shrinivas
Reposted by Krishna Shrinivas
Reposted by Krishna Shrinivas
Reposted by Krishna Shrinivas
Alexis Verger 🧬🧫🧪
@alexis-verger.cpesr.fr
· Jul 18

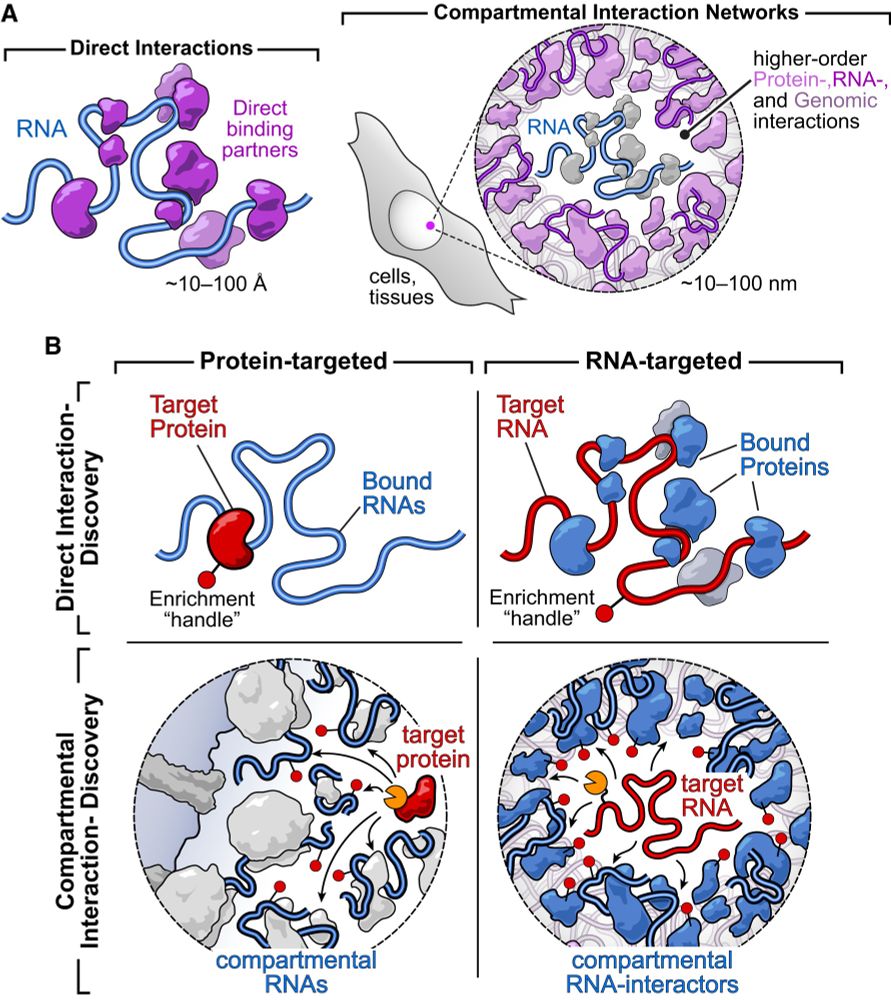
Design of intrinsically disordered region binding proteins
Intrinsically disordered proteins and peptides play key roles in biology, but a lack of defined structures and high variability in sequence and conformational preferences have made targeting such syst...
www.science.org
Reposted by Krishna Shrinivas
Reposted by Krishna Shrinivas
Deepa Rajan
@deeparajan.bsky.social
· Jun 24
Reposted by Krishna Shrinivas
Kalli Kappel
@kallikappel.bsky.social
· Jun 17
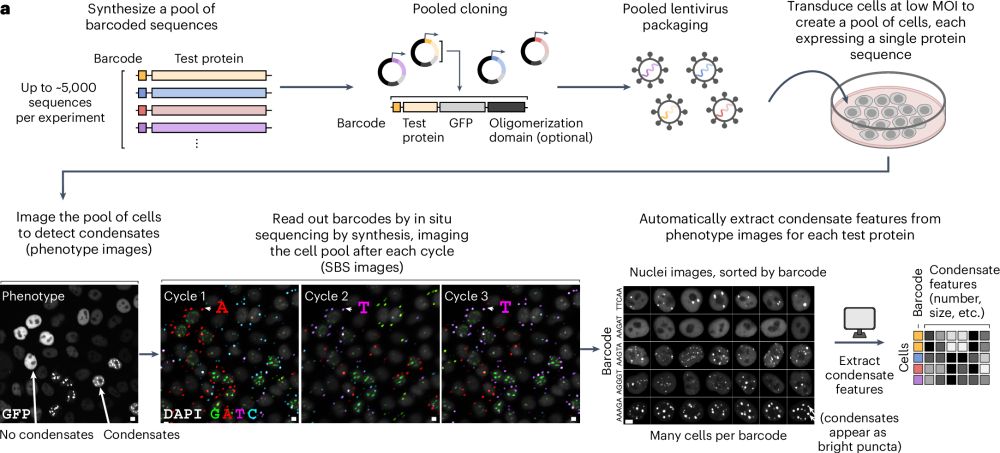
Characterizing protein sequence determinants of nuclear condensates by high-throughput pooled imaging with CondenSeq
Nature Methods - CondenSeq is an imaging-based, high-throughput platform for characterizing condensate formation within the nuclear environment, uncovering the protein sequence features that...
rdcu.be
Reposted by Krishna Shrinivas