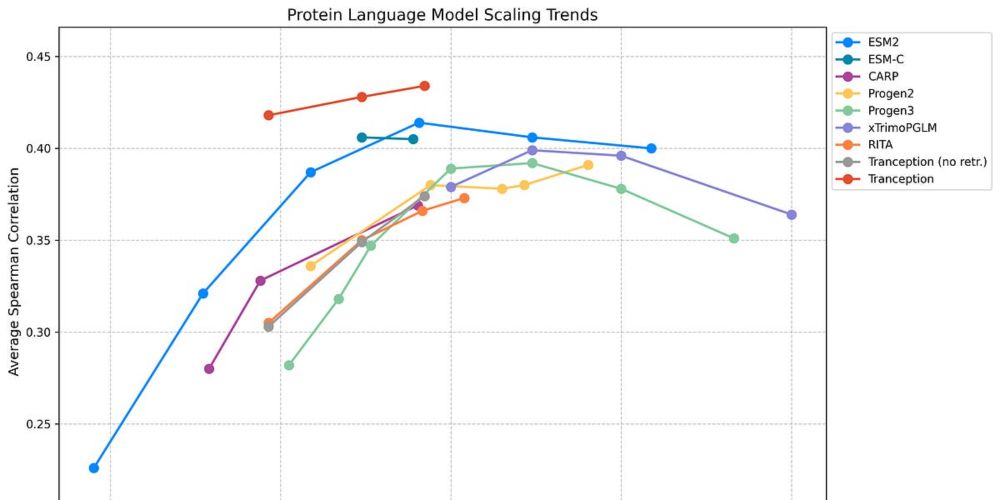
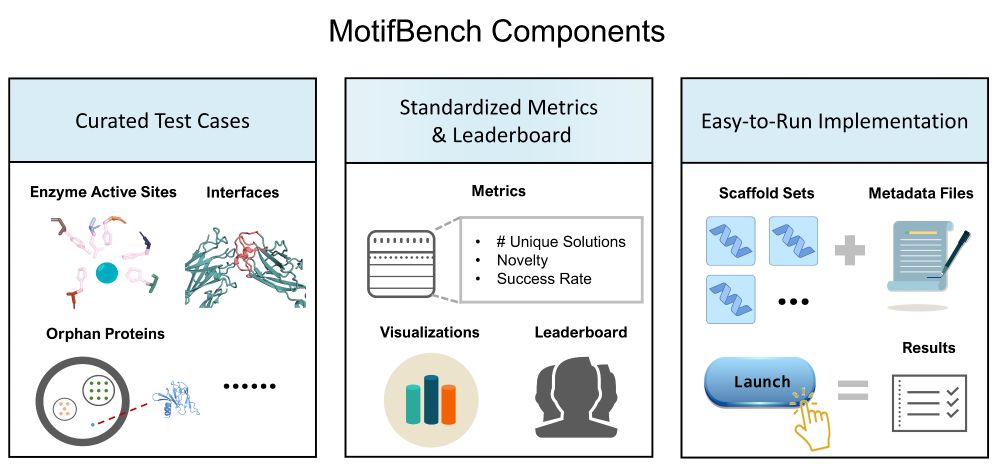
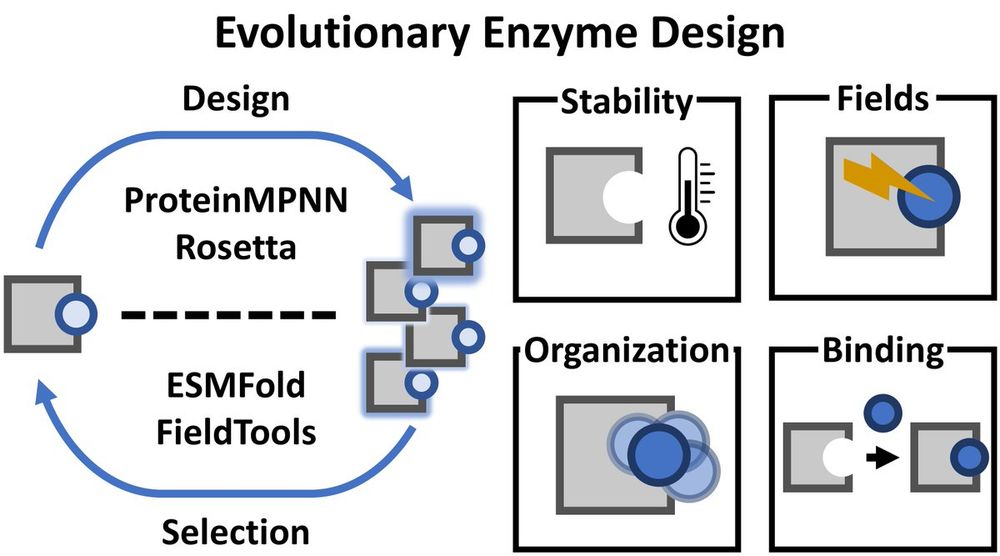
Simon Mathis
@simonmathis.bsky.social
910 followers
580 following
31 posts
PhD student at Uni of Cambridge, UK 🔬 | AI for protein design & engineering 🧬 | biotech & environmental applications 🌱 | enzymes 🏗️
🇦🇹🇨🇭
Posts
Media
Videos
Starter Packs
Pinned
Simon Mathis
@simonmathis.bsky.social
· Nov 20
Reposted by Simon Mathis
Nate Corley
@ncorley.bsky.social
· Aug 15

Accelerating Biomolecular Modeling with AtomWorks and RF3
Deep learning methods trained on protein structure databases have revolutionized biomolecular structure prediction, but developing and training new models remains a considerable challenge. To facilita...
www.biorxiv.org
Reposted by Simon Mathis
Reposted by Simon Mathis
Reposted by Simon Mathis
Reposted by Simon Mathis
Simon Mathis
@simonmathis.bsky.social
· Dec 23
Reposted by Simon Mathis
Gabriele Corso
@gcorso.bsky.social
· Dec 8
Simon Mathis
@simonmathis.bsky.social
· Dec 4
Simon Mathis
@simonmathis.bsky.social
· Dec 2
Simon Mathis
@simonmathis.bsky.social
· Dec 2
Reposted by Simon Mathis
Simon Mathis
@simonmathis.bsky.social
· Nov 30
Simon Mathis
@simonmathis.bsky.social
· Nov 29
Reposted by Simon Mathis
Reposted by Simon Mathis
Simon Mathis
@simonmathis.bsky.social
· Nov 26
Simon Mathis
@simonmathis.bsky.social
· Nov 26
Simon Mathis
@simonmathis.bsky.social
· Nov 26
Simon Mathis
@simonmathis.bsky.social
· Nov 25
GitHub - Croydon-Brixton/pymol-remote: Send data to and from pymol from a remote server (e.g. a cluster running deep learning workflows)
Send data to and from pymol from a remote server (e.g. a cluster running deep learning workflows) - Croydon-Brixton/pymol-remote
github.com
Simon Mathis
@simonmathis.bsky.social
· Nov 25