Sandra Segura-Bayona
@ssegurabayona.bsky.social
420 followers
470 following
9 posts
Postdoctoral researcher in the Boulton Lab 🧬 The Francis Crick Institute, London, UK▪️interest in genome stability, chromatin, cancer 🧫 👩🔬 EMBO and MSCA fellow
Posts
Media
Videos
Starter Packs
Reposted by Sandra Segura-Bayona
Reposted by Sandra Segura-Bayona
Reposted by Sandra Segura-Bayona
Reposted by Sandra Segura-Bayona
Reposted by Sandra Segura-Bayona
Reposted by Sandra Segura-Bayona
Reposted by Sandra Segura-Bayona
Reposted by Sandra Segura-Bayona
Reposted by Sandra Segura-Bayona
Dirk Remus
@dirkremus.bsky.social
· Mar 6
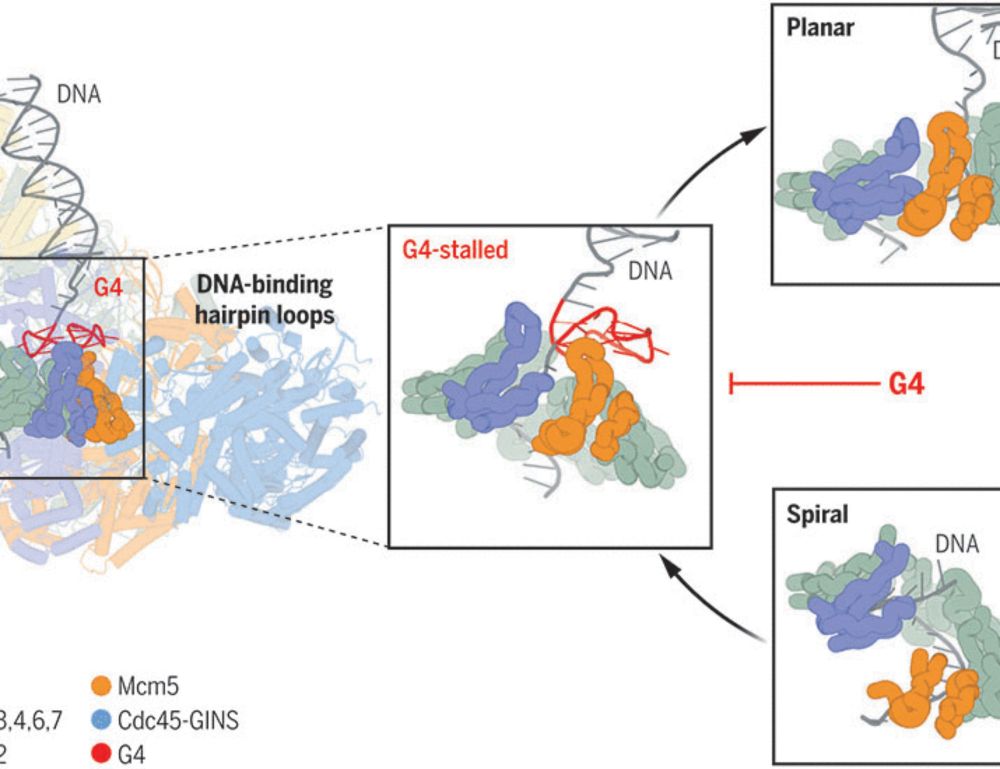
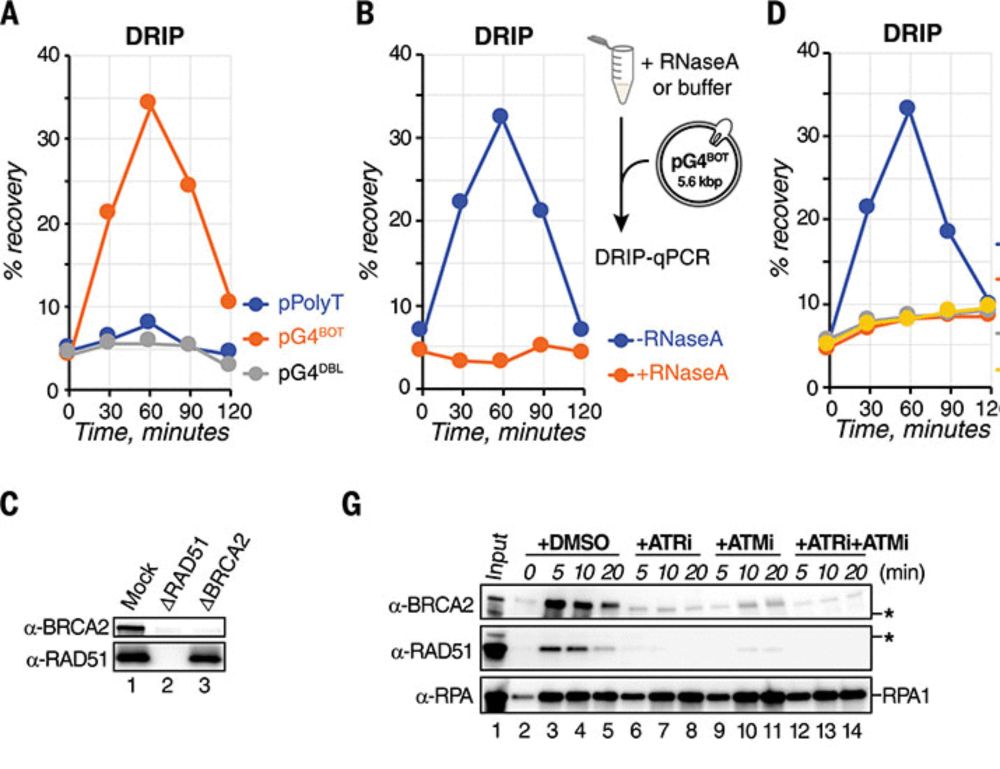
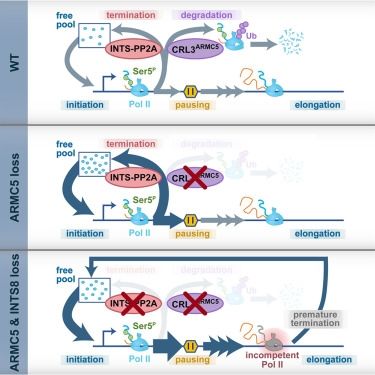
G-quadruplex–stalled eukaryotic replisome structure reveals helical inchworm DNA translocation
DNA G-quadruplexes (G4s) are non–B-form DNA secondary structures that threaten genome stability by impeding DNA replication. To elucidate how G4s induce replication fork arrest, we characterized fork ...
www.science.org
Reposted by Sandra Segura-Bayona
The Groth lab
@grothlab.bsky.social
· Feb 19
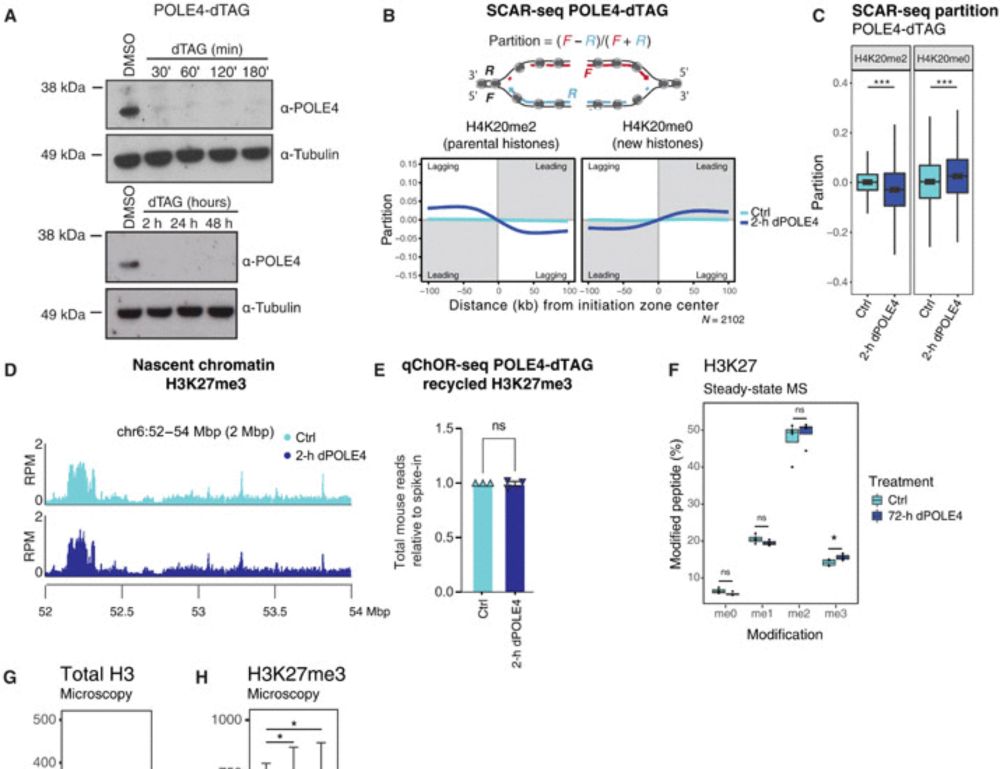
Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability
Losing parental histones during DNA replication fork passage challenges differentiation competence and cell viability.
tinyurl.com
Reposted by Sandra Segura-Bayona
Tony Cesare
@thecesarelab.bsky.social
· Jan 13
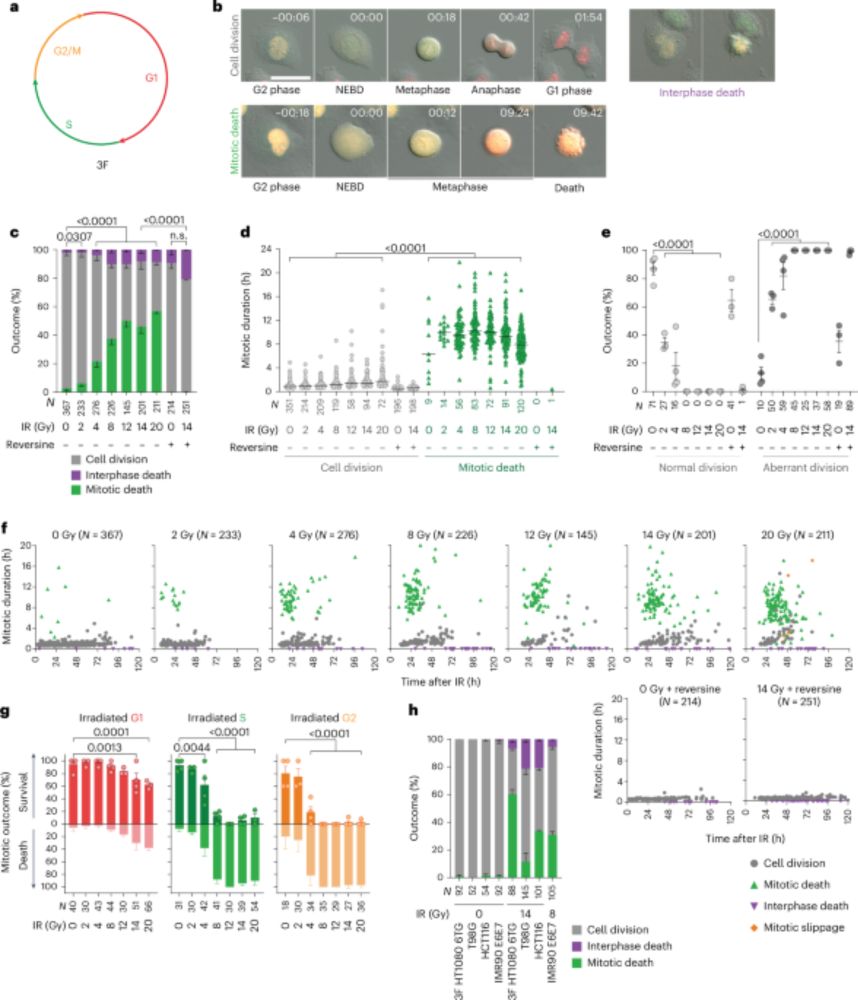
Homologous recombination promotes non-immunogenic mitotic cell death upon DNA damage - Nature Cell Biology
Szmyd et al. show that DNA repair pathways impact whether cells with DNA lesions arrest in mitosis. The formation of homologous recombination-driven double Holliday junctions elicits mitotic cell deat...
www.nature.com
Reposted by Sandra Segura-Bayona