Surag Nair
@suragnair.bsky.social
660 followers
460 following
5 posts
Machine learning and genetics @Genentech. Previously CS PhD @Stanford.
suragnair.github.io
Posts
Media
Videos
Starter Packs
Surag Nair
@suragnair.bsky.social
· Mar 19
Reposted by Surag Nair
Peter Koo
@pkoo562.bsky.social
· Feb 5
Surag Nair
@suragnair.bsky.social
· Dec 22
Surag Nair
@suragnair.bsky.social
· Dec 5
2025 Summer Intern - Biology Research | AI Development
2025 Summer Intern - Biology Research | AI Development Department Summary At Genentech Research & Early Development (gRED) we have initiated an exciting journey to bring together and further stren...
roche.wd3.myworkdayjobs.com
Reposted by Surag Nair
Reposted by Surag Nair
Mitch Guttman
@mitchguttman.bsky.social
· Nov 27
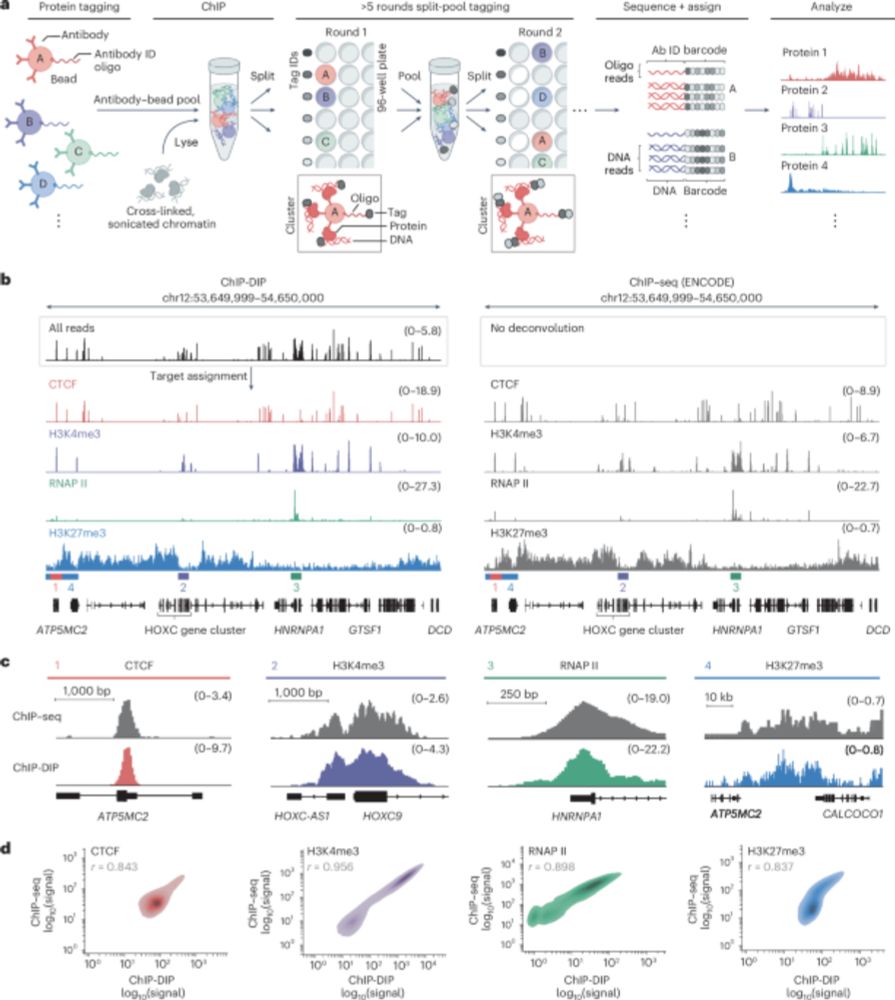
ChIP-DIP maps binding of hundreds of proteins to DNA simultaneously and identifies diverse gene regulatory elements - Nature Genetics
ChIP-DIP (ChIP done in parallel) is a highly multiplex assay for protein–DNA binding, scalable to hundreds of proteins including modified histones, chromatin regulators and transcription factors, offe...
www.nature.com
Reposted by Surag Nair
Wei-Lin Qiu
@613weilin.bsky.social
· Nov 25

Mapping enhancer-gene regulatory interactions from single-cell data
Mapping enhancers and their target genes in specific cell types is crucial for understanding gene regulation and human disease genetics. However, accurately predicting enhancer-gene regulatory interac...
biorxiv.org
Reposted by Surag Nair
Alex Thiery
@alexxthiery.bsky.social
· Nov 23
Reposted by Surag Nair
Dan Roy
@roydanroy.bsky.social
· Nov 22
Reposted by Surag Nair
Reposted by Surag Nair
Jesse Engreitz
@jengreitz.bsky.social
· Nov 20
Reposted by Surag Nair
Reposted by Surag Nair
Pooja Kathail
@poojakathail.bsky.social
· Nov 20

Leveraging genomic deep learning models for non-coding variant effect prediction
The majority of genetic variants identified in genome-wide association studies of complex traits are non-coding, and characterizing their function remains an important challenge in human genetics. Gen...
arxiv.org
Reposted by Surag Nair
Shaun Mahony
@shaunmahony.bsky.social
· Nov 18
Surag Nair
@suragnair.bsky.social
· Nov 19
Reposted by Surag Nair
Reposted by Surag Nair
Shaun Mahony
@shaunmahony.bsky.social
· Nov 14
Reposted by Surag Nair
Anshul Kundaje
@anshulkundaje.bsky.social
· Nov 13

Extensive binding of uncharacterized human transcription factors to genomic dark matter
Most of the human genome is thought to be non-functional, and includes large segments often referred to as "dark matter" DNA. The genome also encodes hundreds of putative and poorly characterized tran...
www.biorxiv.org

