
www.nature.com/articles/s41...
Led by @bendavisgroup.bsky.social and with some very deep chemoproteomics characterisation

www.nature.com/articles/s41...
Led by @bendavisgroup.bsky.social and with some very deep chemoproteomics characterisation
www.biorxiv.org/content/10.6...

Cross-species ML: M.tb compound-protein X-attn & human chemoproteomics.




Cross-species ML: M.tb compound-protein X-attn & human chemoproteomics.
pubs.acs.org/doi/full/10....
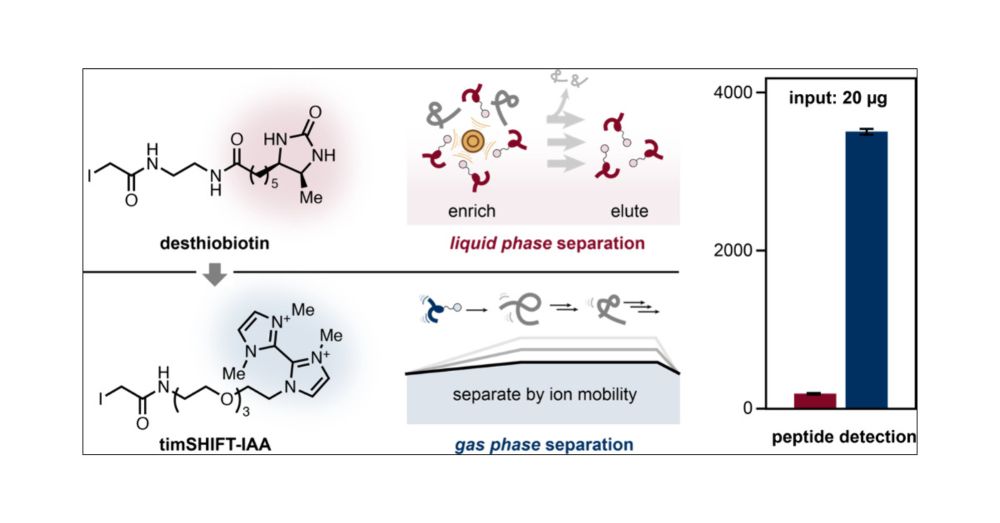
pubs.acs.org/doi/full/10....

Check it out in full here🔽


Like Mork & Mindy, these stinky residues love to bond! Jerks like us break these bonds, with chemoproteomics trying to map, probe, and manipulate them. Leftovers stuck to S-S is a fun field but I love them for their antioxidant and metal binding abilities.

Like Mork & Mindy, these stinky residues love to bond! Jerks like us break these bonds, with chemoproteomics trying to map, probe, and manipulate them. Leftovers stuck to S-S is a fun field but I love them for their antioxidant and metal binding abilities.

#ChemSky #ChemBio #ProudPI

#ChemSky #ChemBio #ProudPI
#Chembio #Proteomics #chemoproteomics

#Chembio #Proteomics #chemoproteomics
If you want to check it out, you can find it here: www.nature.com/articles/s41...
#ChemSky #ChemBio #ChemicalProteomics #Chemoproteomics #ProteoProbes #ChemPro

If you want to check it out, you can find it here: www.nature.com/articles/s41...
#ChemSky #ChemBio #ChemicalProteomics #Chemoproteomics #ProteoProbes #ChemPro
Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...

Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...

Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social
Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...

Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social
"These cases...point to the need to re-train ML co-folding tools on datasets enriched in more diverse types of small molecule-protein interactions...to avoid biased predictions derived from memorization of common orthosteric ligand poses"
www.biorxiv.org/content/10.1...

"These cases...point to the need to re-train ML co-folding tools on datasets enriched in more diverse types of small molecule-protein interactions...to avoid biased predictions derived from memorization of common orthosteric ligand poses"
www.biorxiv.org/content/10.1...
Tryptoline stereoprobes reveal hidden cysteine ligandability: while Boltz-2 nails orthosteric sites but burns out on non-orthosteric ones.
A masterpiece of chemoproteomics + AI.
🔗 doi.org/10.1101/2025...

Tryptoline stereoprobes reveal hidden cysteine ligandability: while Boltz-2 nails orthosteric sites but burns out on non-orthosteric ones.
A masterpiece of chemoproteomics + AI.
🔗 doi.org/10.1101/2025...
SuPUR revealed an ACAT2-Y237 ligand uncovering a cancer metabolic vulnerability.
www.biorxiv.org/content/10.1...

SuPUR revealed an ACAT2-Y237 ligand uncovering a cancer metabolic vulnerability.
www.biorxiv.org/content/10.1...
Chemoproteomics were used to uncover a previously unknown serine protease Bfp1 from B fragilis! This sneaky enzyme cleaves PAR2, setting off a chain reaction of barrier dysfunction, colonic inflammation, and nociceptor activation
pubmed.ncbi.nlm.nih.gov/41015045/

Chemoproteomics were used to uncover a previously unknown serine protease Bfp1 from B fragilis! This sneaky enzyme cleaves PAR2, setting off a chain reaction of barrier dysfunction, colonic inflammation, and nociceptor activation
pubmed.ncbi.nlm.nih.gov/41015045/
"chemoproteomics"
"chemoproteomics"
www.nature.com/articles/s41...

www.nature.com/articles/s41...

