
www.nature.com/articles/s41...
A fun collaboration with @beckmannlab.bsky.social @doubleshuang.bsky.social

www.nature.com/articles/s41...
A fun collaboration with @beckmannlab.bsky.social @doubleshuang.bsky.social
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Or from a single dish of HEK cell culture in the case of two membrane proteins.
Out in Nature Methods now! lnkd.in/gpyBSceg
Wonderful collaboration with the Efremov lab.

Or from a single dish of HEK cell culture in the case of two membrane proteins.
Out in Nature Methods now! lnkd.in/gpyBSceg
Wonderful collaboration with the Efremov lab.



www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
New preprint!
How do transcription factors (TFs) use intrinsically disordered regions (IDRs) to find their target sites?
www.biorxiv.org/content/10.1...
#TranscriptionFactors #IDPs #SingleMolecule #Biophysics

New preprint!
How do transcription factors (TFs) use intrinsically disordered regions (IDRs) to find their target sites?
www.biorxiv.org/content/10.1...
#TranscriptionFactors #IDPs #SingleMolecule #Biophysics
@natsmb.nature.com
1/7
www.nature.com/articles/s41...
@natsmb.nature.com
1/7
www.nature.com/articles/s41...
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
www.nature.com/articles/s41...
www.nature.com/articles/s41...
pmc.ncbi.nlm.nih.gov/articles/PMC...

pmc.ncbi.nlm.nih.gov/articles/PMC...
You'll find a tutorial on how to reconstruct tomograms, pick particles and do subtomogram averaging, using different software!
Hope it will be useful !


You'll find a tutorial on how to reconstruct tomograms, pick particles and do subtomogram averaging, using different software!
Hope it will be useful !
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.nature.com/articles/s42...
www.nature.com/articles/s42...
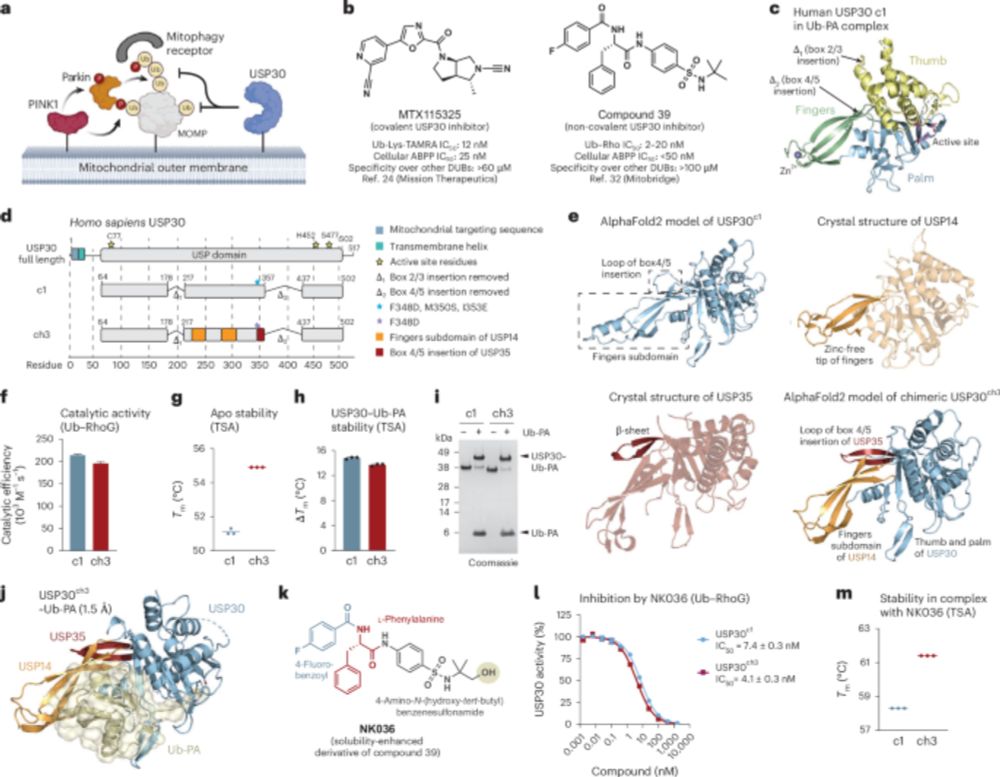

www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
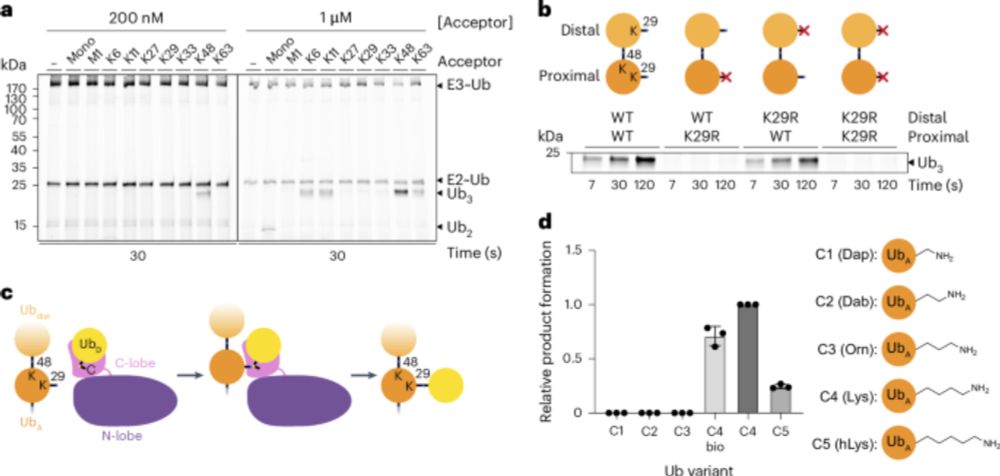
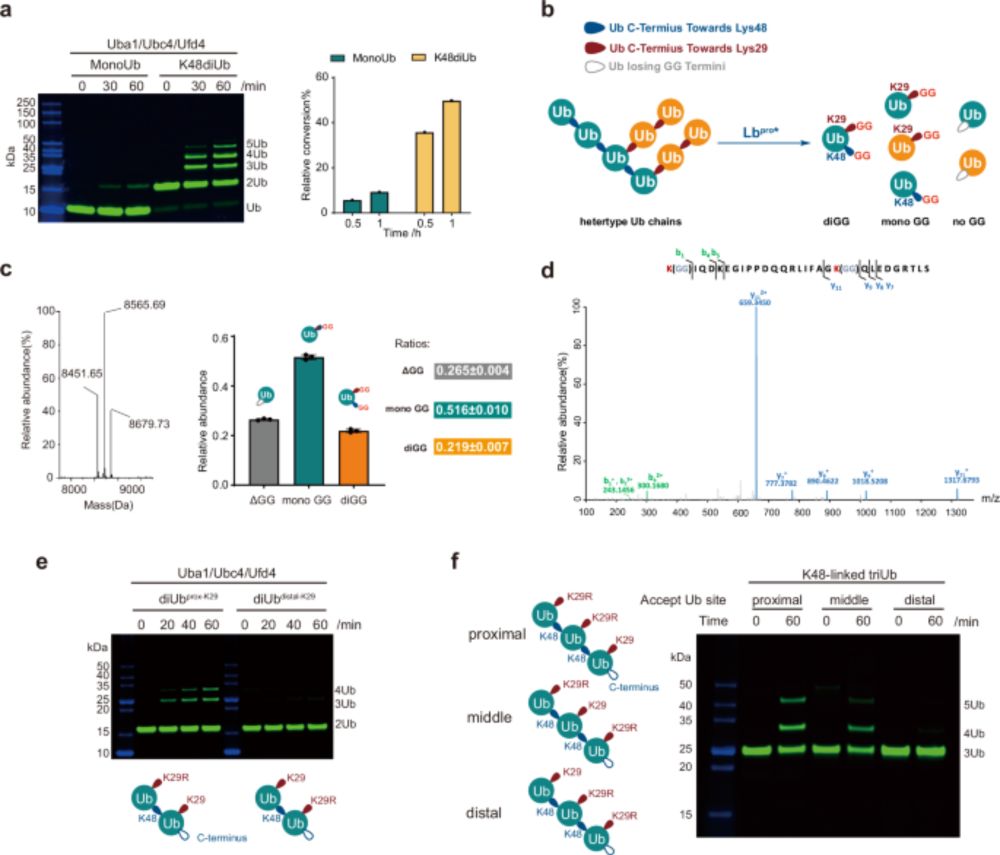
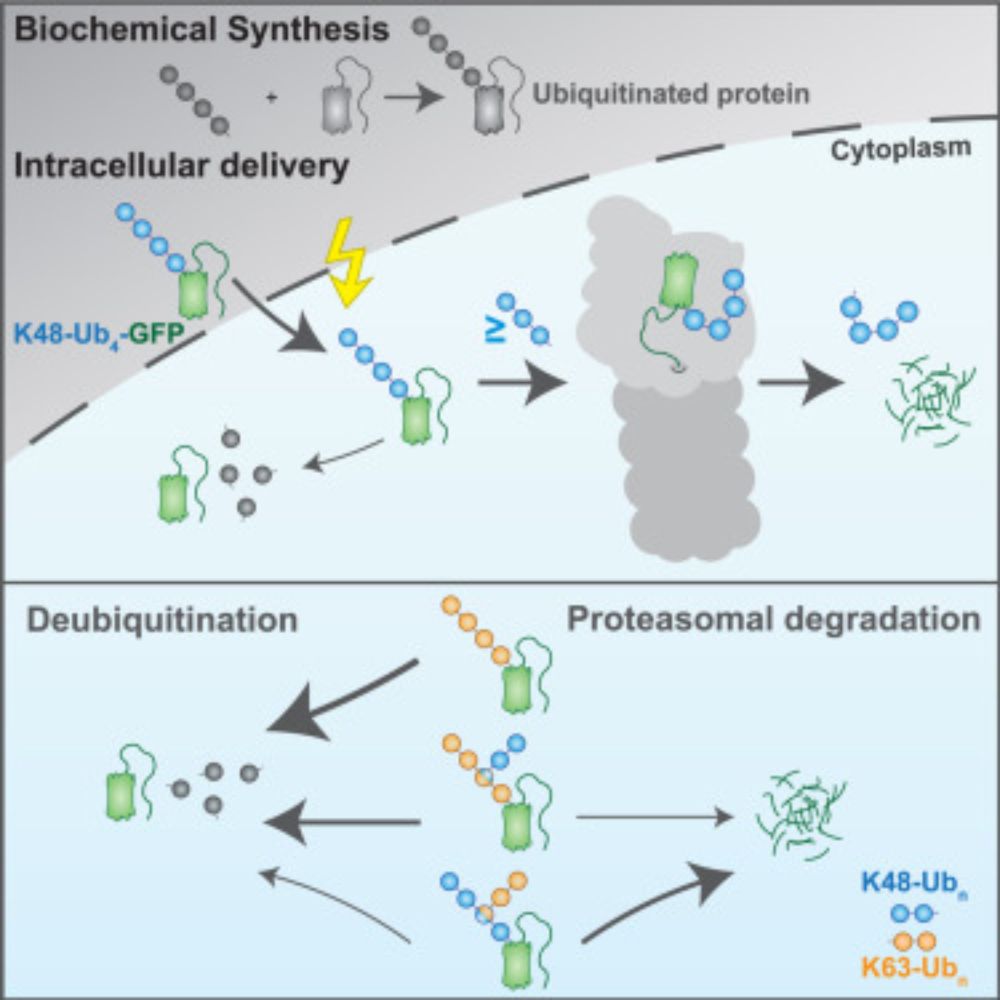
“𝗨𝗯𝗶𝗥𝗘𝗔𝗗 𝗱𝗲𝗰𝗶𝗽𝗵𝗲𝗿𝘀 𝗽𝗿𝗼𝘁𝗲𝗮𝘀𝗼𝗺𝗮𝗹 𝗱𝗲𝗴𝗿𝗮𝗱𝗮𝘁𝗶𝗼𝗻 𝗰𝗼𝗱𝗲 𝗼𝗳 𝗵𝗼𝗺𝗼𝘁𝘆𝗽𝗶𝗰 𝗮𝗻𝗱 𝗯𝗿𝗮𝗻𝗰𝗵𝗲𝗱 𝗞48 𝗮𝗻𝗱 𝗞63 𝘂𝗯𝗶𝗾𝘂𝗶𝘁𝗶𝗻 𝗰𝗵𝗮𝗶𝗻𝘀”
@cp-molcell.bsky.social
1/8
www.cell.com/molecular-ce...
“𝗨𝗯𝗶𝗥𝗘𝗔𝗗 𝗱𝗲𝗰𝗶𝗽𝗵𝗲𝗿𝘀 𝗽𝗿𝗼𝘁𝗲𝗮𝘀𝗼𝗺𝗮𝗹 𝗱𝗲𝗴𝗿𝗮𝗱𝗮𝘁𝗶𝗼𝗻 𝗰𝗼𝗱𝗲 𝗼𝗳 𝗵𝗼𝗺𝗼𝘁𝘆𝗽𝗶𝗰 𝗮𝗻𝗱 𝗯𝗿𝗮𝗻𝗰𝗵𝗲𝗱 𝗞48 𝗮𝗻𝗱 𝗞63 𝘂𝗯𝗶𝗾𝘂𝗶𝘁𝗶𝗻 𝗰𝗵𝗮𝗶𝗻𝘀”
@cp-molcell.bsky.social
1/8
www.cell.com/molecular-ce...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
www.nature.com/articles/s41...
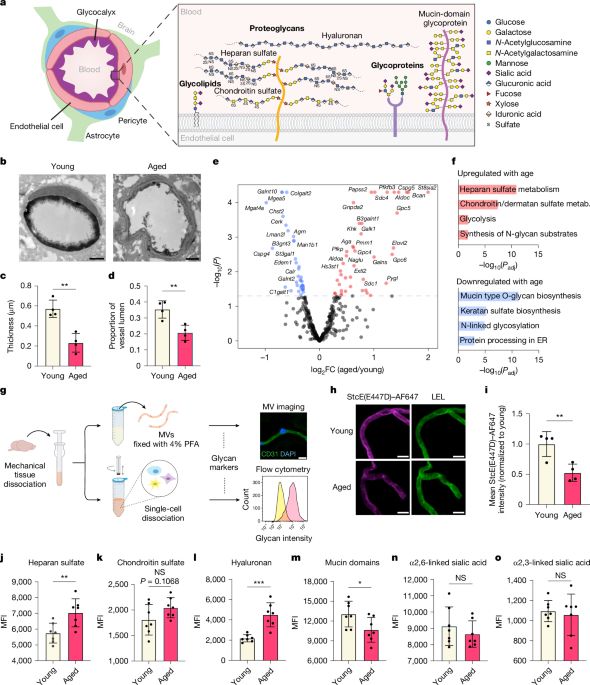
www.nature.com/articles/s41...


