

Many thanks to @beilstein-institut.bsky.social @glycosmos.bsky.social @sib.swiss

Many thanks to @beilstein-institut.bsky.social @glycosmos.bsky.social @sib.swiss

Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social
Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...

Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social



openrxiv.org/enabling-rev...

Thanks to @willfondrie.com for alerting me to this amazing repo.
genzplyr is dplyr, but bussin fr fr no cap.

Thanks to @willfondrie.com for alerting me to this amazing repo.

A collaboration to give the Flaviviridae (home to Zika, Dengue & HCV) a much-needed taxonomic re-think.
Our at-scale AI structure prediction gave a complementary perspective on viral evolution.
www.nature.com/articles/s41...

A collaboration to give the Flaviviridae (home to Zika, Dengue & HCV) a much-needed taxonomic re-think.
Our at-scale AI structure prediction gave a complementary perspective on viral evolution.
www.nature.com/articles/s41...
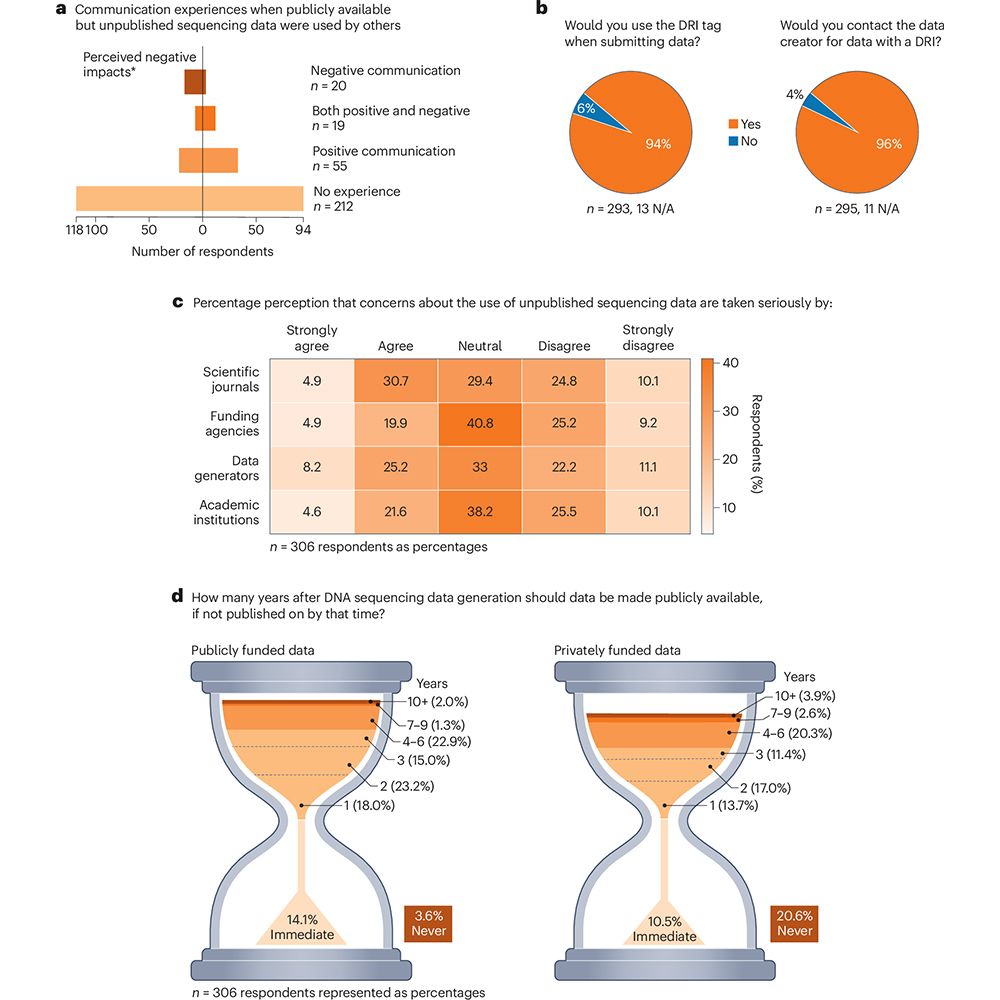
Read all about it in our preprint by👉 doi.org/10.1101/2025...
#Proteomics #AI #MachineLearning

Read all about it in our preprint by👉 doi.org/10.1101/2025...
#Proteomics #AI #MachineLearning
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...
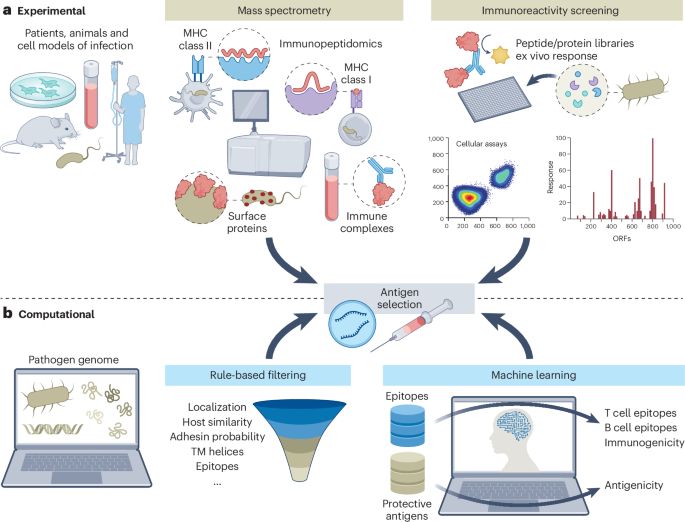
www.nature.com/articles/s41...

---
#proteomics #prot-paper

---
#proteomics #prot-paper
Take advantage of this extra time to share your latest proteomics discoveries.
Submit now: www.ushupoconference.org

Take advantage of this extra time to share your latest proteomics discoveries.
Submit now: www.ushupoconference.org
---
#proteomics #prot-paper
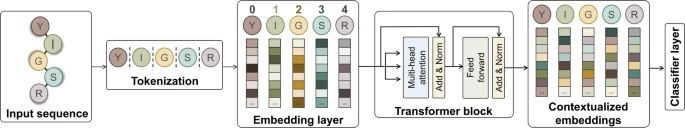
---
#proteomics #prot-paper
---
#proteomics #prot-paper

---
#proteomics #prot-paper




• Especially useful for preclinical metabolites, that have no standards.
Analytical Chemistry (open access article)
pubs.acs.org/doi/10.1021/...
• Especially useful for preclinical metabolites, that have no standards.
Analytical Chemistry (open access article)
pubs.acs.org/doi/10.1021/...

