
#teamTomo
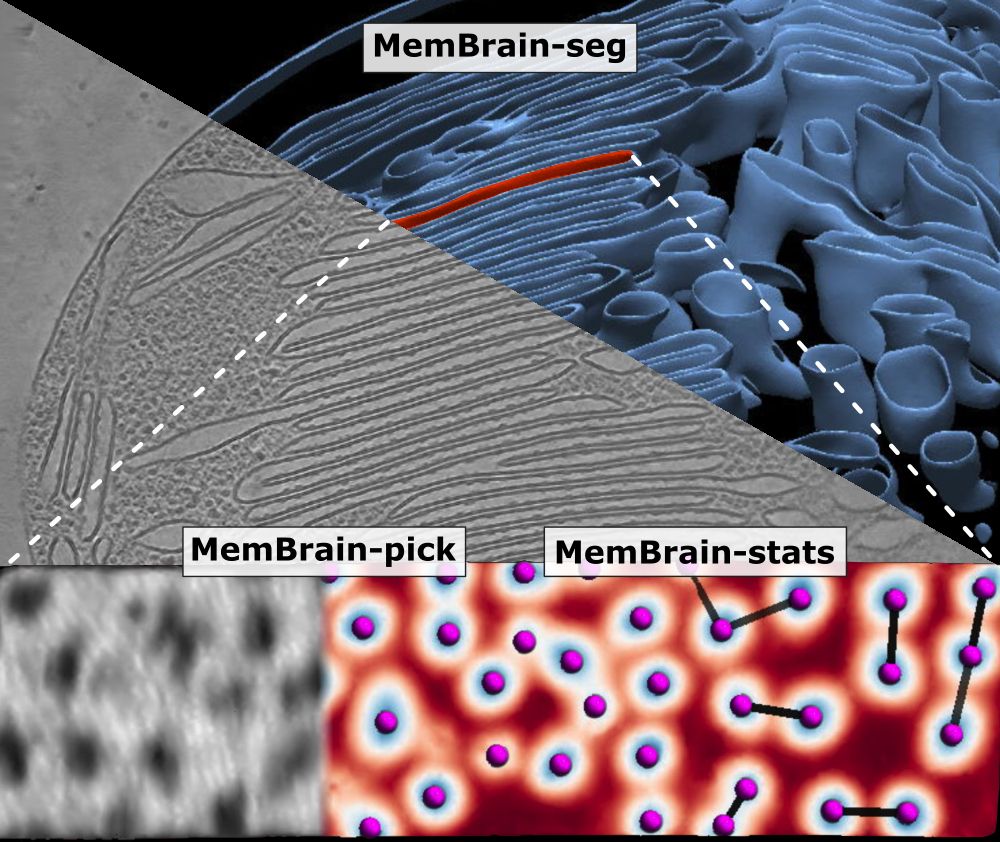
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
Preprint here!:
www.biorxiv.org/content/10.6...
@cellarchlab.com @phaips.vd.st @biologyatyork.bsky.social
#Rubisco #PhaseSeparation #Condensates #Pyrenoid
Preprint here!:
www.biorxiv.org/content/10.6...
@cellarchlab.com @phaips.vd.st @biologyatyork.bsky.social
#Rubisco #PhaseSeparation #Condensates #Pyrenoid
This beautiful rendering made by co-author @jessheebner.bsky.social and Holly Peterson shows an instance of mitochondrial fission found in the dataset 😍
[Maybe long thread ahead]
This beautiful rendering made by co-author @jessheebner.bsky.social and Holly Peterson shows an instance of mitochondrial fission found in the dataset 😍
[Maybe long thread ahead]
Read @lifeonthewedge.bsky.social's great thread🧵 for the inside scoop🍨 on all the #TeamTomo developments already made possible since these 1829 tomos hit EMPIAR 🧪 🧶🧬
For more, here's the old preprint thread:
bsky.app/profile/cell...
This beautiful rendering made by co-author @jessheebner.bsky.social and Holly Peterson shows an instance of mitochondrial fission found in the dataset 😍
[Maybe long thread ahead]
Read @lifeonthewedge.bsky.social's great thread🧵 for the inside scoop🍨 on all the #TeamTomo developments already made possible since these 1829 tomos hit EMPIAR 🧪 🧶🧬
For more, here's the old preprint thread:
bsky.app/profile/cell...

🔗 Preprint: doi.org/10.48550/arX...
🔗 Code: github.com/peng-lab/rmlp
🧵1/9

🔗 Preprint: doi.org/10.48550/arX...
🔗 Code: github.com/peng-lab/rmlp
🧵1/9
@virtualhomo.bsky.social found an elegant way to regularize DINOv2 training using randomised linear algebra 🤯
Check out his thread or even his poster if you're at #NeurIPS2025.

@virtualhomo.bsky.social found an elegant way to regularize DINOv2 training using randomised linear algebra 🤯
Check out his thread or even his poster if you're at #NeurIPS2025.
So if u fit there, u might wanna check it out! Maybe also if you don't. We're allies here <3

So if u fit there, u might wanna check it out! Maybe also if you don't. We're allies here <3
New features include:
* Training on multiple tomograms (same training time, with linear increase in RAM) 🚀
* Logging and plotting of the loss function 📉
* The --scale option is now called --eq-weight for clarity 😉
We'd love to hear your feedback! 🙏🏽
New features include:
* Training on multiple tomograms (same training time, with linear increase in RAM) 🚀
* Logging and plotting of the loss function 📉
* The --scale option is now called --eq-weight for clarity 😉
We'd love to hear your feedback! 🙏🏽
napari.org/island-dispa...
napari.org/island-dispa...
Here, we used #CryoET to visualise mitochondrial proteostatic stress and together with SPA #CryoEM shed light into the functional cycle of the Hsp60:10 chaperone system. #TeamTomo
🔗 www.biorxiv.org/content/10.1...

Here, we used #CryoET to visualise mitochondrial proteostatic stress and together with SPA #CryoEM shed light into the functional cycle of the Hsp60:10 chaperone system. #TeamTomo
🔗 www.biorxiv.org/content/10.1...
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
buff.ly/j3TSIkn
buff.ly/j3TSIkn
We built a phantom cryo-ET dataset (~500 tomograms) + hosted a Kaggle challenge.
The result: community models beat expert tools.
Read more in the @natmethods.nature.com article that just came out:
🔗 doi.org/10.1038/s415...

We built a phantom cryo-ET dataset (~500 tomograms) + hosted a Kaggle challenge.
The result: community models beat expert tools.
Read more in the @natmethods.nature.com article that just came out:
🔗 doi.org/10.1038/s415...
Through the dedication of @glynnca.bsky.social and @cryingem.bsky.social we report a thorough method to image molecular organisation within hippocampus tissue.
Structural biology in tissue is well and truly here!
@rosfrankinst.bsky.social
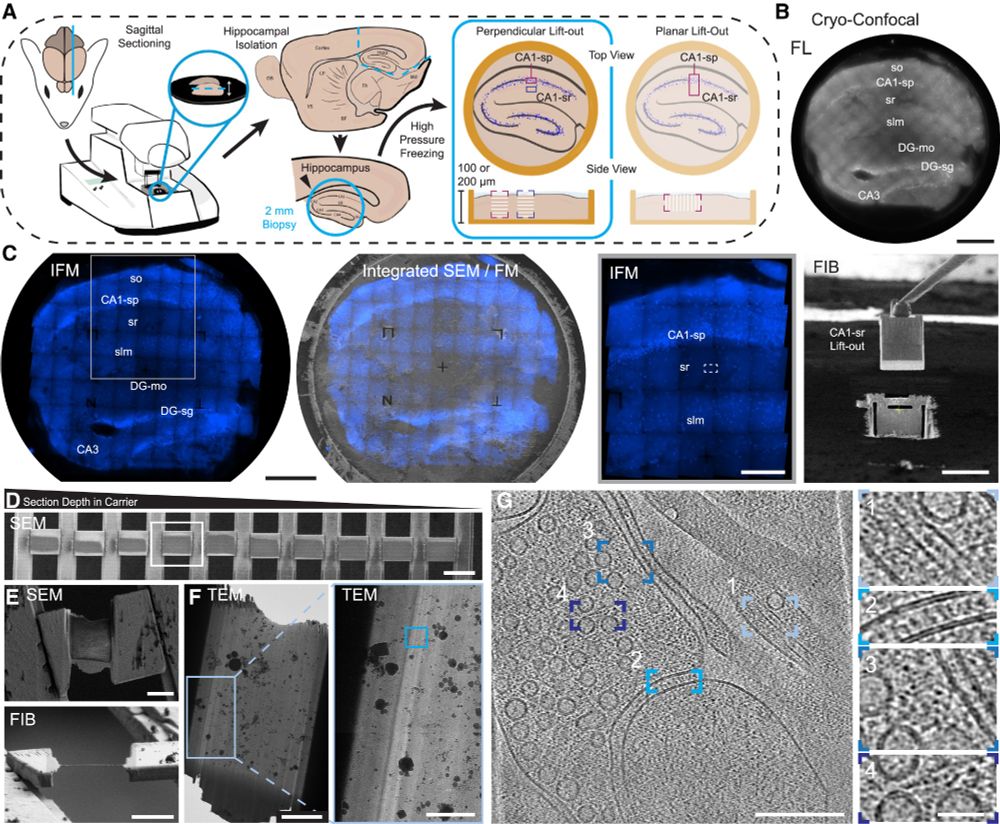
Through the dedication of @glynnca.bsky.social and @cryingem.bsky.social we report a thorough method to image molecular organisation within hippocampus tissue.
Structural biology in tissue is well and truly here!
@rosfrankinst.bsky.social
📅 1 July 9AM CEST
🔗 tinyurl.com/4mjaverj
Spread the word!
@protistwtmostest.bsky.social @ehehenberger.bsky.social @chandnibhickta.bsky.social&Norico Yamada

📅 1 July 9AM CEST
🔗 tinyurl.com/4mjaverj
Spread the word!
@protistwtmostest.bsky.social @ehehenberger.bsky.social @chandnibhickta.bsky.social&Norico Yamada
You'll find a tutorial on how to reconstruct tomograms, pick particles and do subtomogram averaging, using different software!
Hope it will be useful !


You'll find a tutorial on how to reconstruct tomograms, pick particles and do subtomogram averaging, using different software!
Hope it will be useful !
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
paper: www.biorxiv.org/content/10.1...
w/ @computingnature.bsky.social 1/n
paper: www.biorxiv.org/content/10.1...
w/ @computingnature.bsky.social 1/n
Cryo-ET membrane analysis via deep learning.

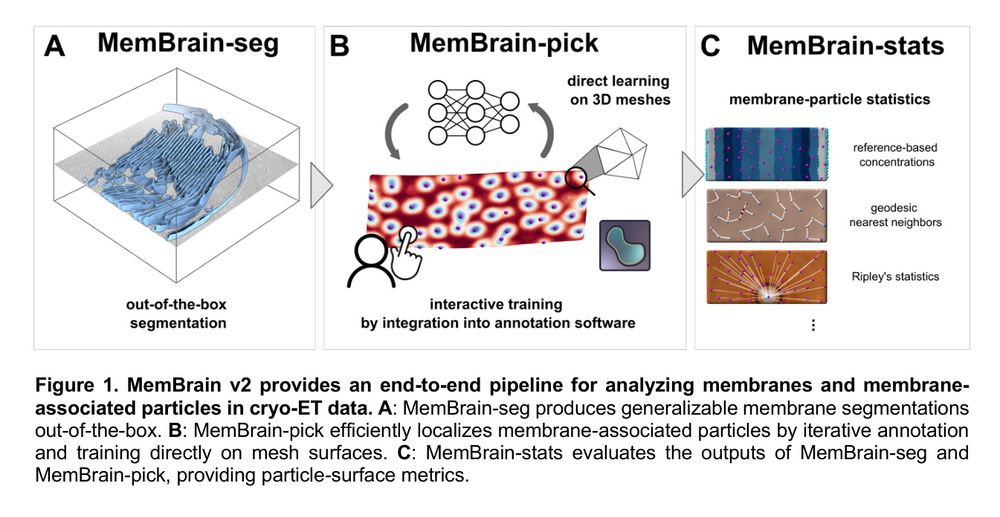
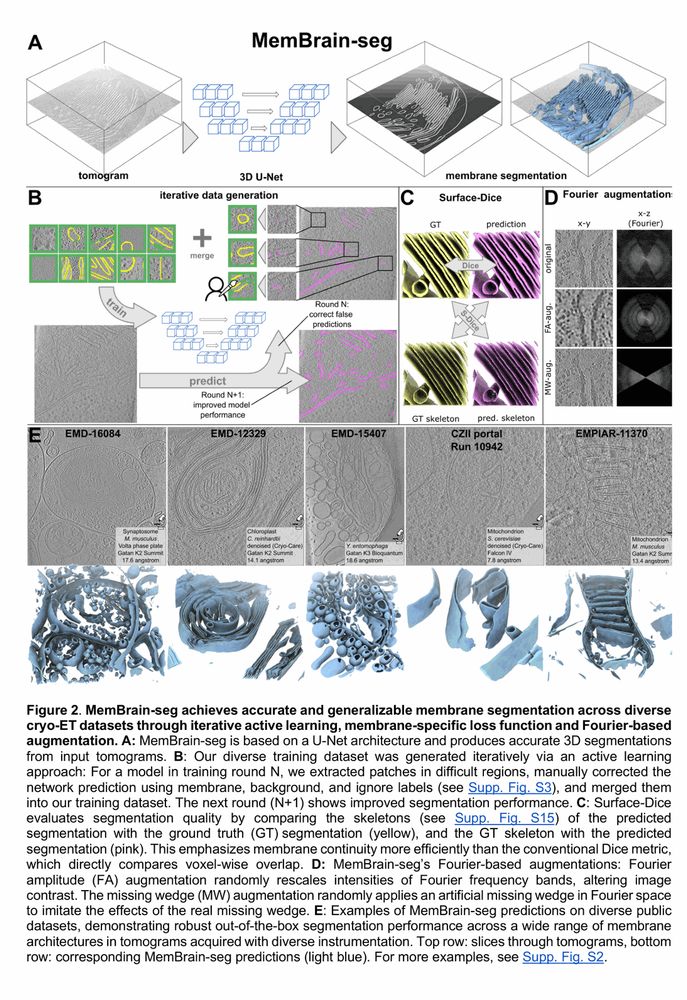
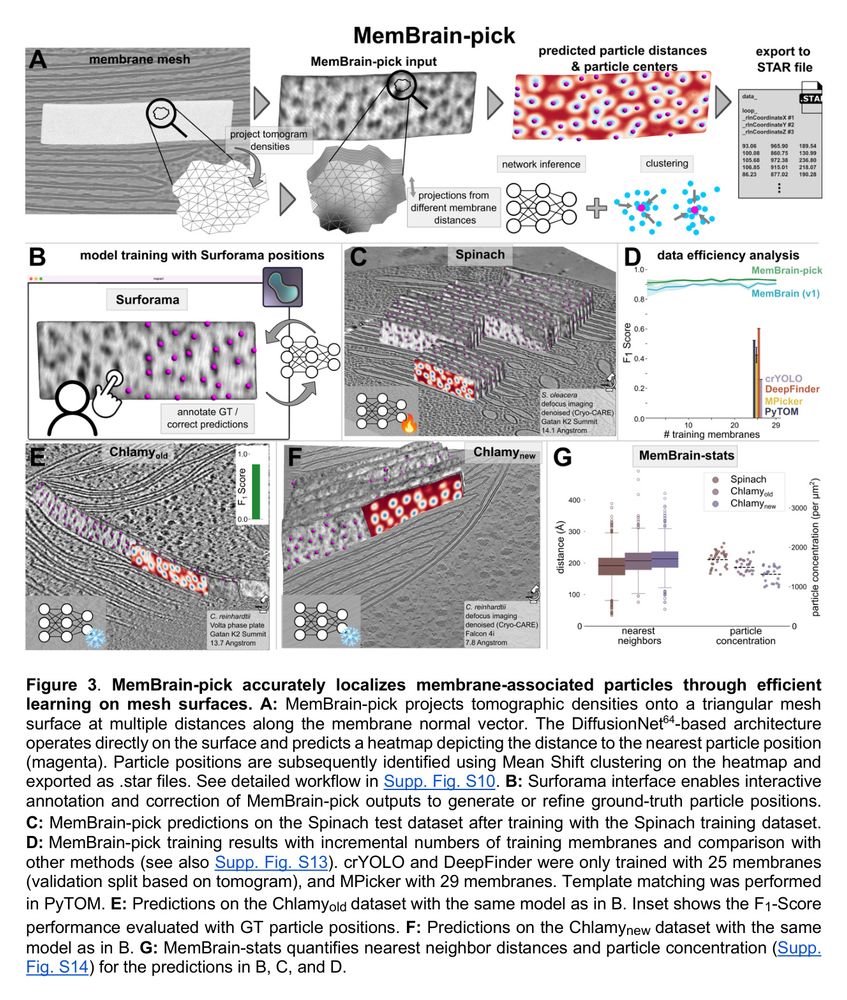
Cryo-ET membrane analysis via deep learning.
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo

We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo

We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo

We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
Stay tuned for the upcoming thread by lead author @lorenzlamm.bsky.social! 🧠🧵
#CryoET #TeamTomo
www.biorxiv.org/content/10.1...
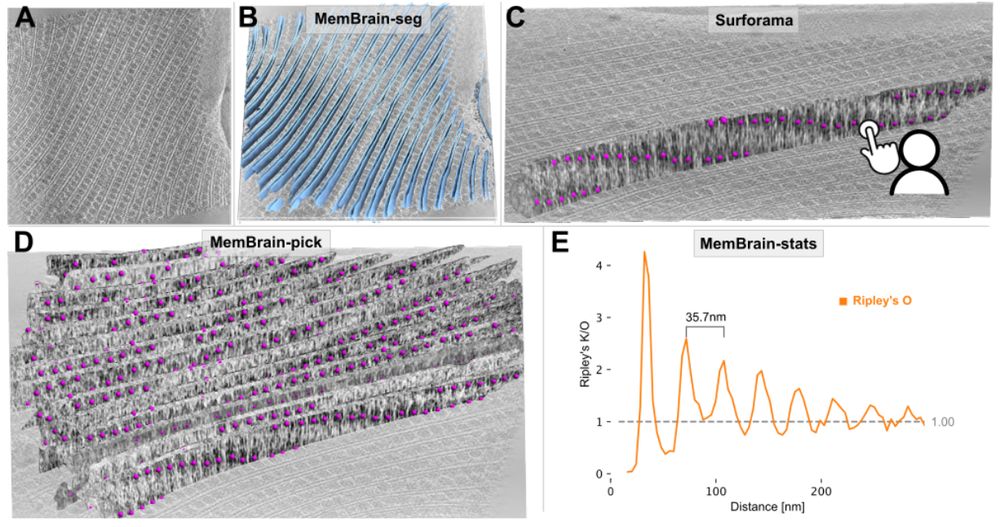
Stay tuned for the upcoming thread by lead author @lorenzlamm.bsky.social! 🧠🧵
#CryoET #TeamTomo
www.biorxiv.org/content/10.1...

Stay tuned for the upcoming thread by lead author @lorenzlamm.bsky.social! 🧠🧵
#CryoET #TeamTomo
www.biorxiv.org/content/10.1...

