Lorenz Lamm
@lorenzlamm.bsky.social
240 followers
300 following
12 posts
PhD Student at Helmholtz AI | MemBrain analysis for Cryo-ET
#teamTomo
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Kyle Harrington
@kyleharrington.com
· Aug 26

A realistic phantom dataset for benchmarking cryo-ET data annotation - Nature Methods
A standardized, realistic phantom dataset consisting of ground-truth annotations for six diverse molecular species is provided as a community resource for cryo-electron-tomography algorithm benchmarki...
doi.org
Reposted by Lorenz Lamm
Lorenz Lamm
@lorenzlamm.bsky.social
· Jun 16
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
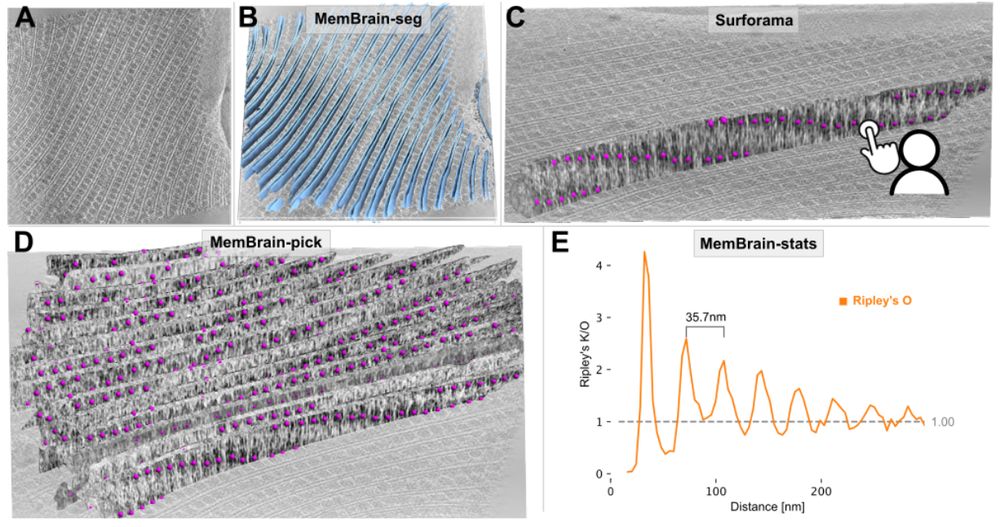
Lorenz Lamm
@lorenzlamm.bsky.social
· Apr 25
Lorenz Lamm
@lorenzlamm.bsky.social
· Apr 25
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Reposted by Lorenz Lamm
Lorenz Lamm
@lorenzlamm.bsky.social
· Apr 8
Reposted by Lorenz Lamm