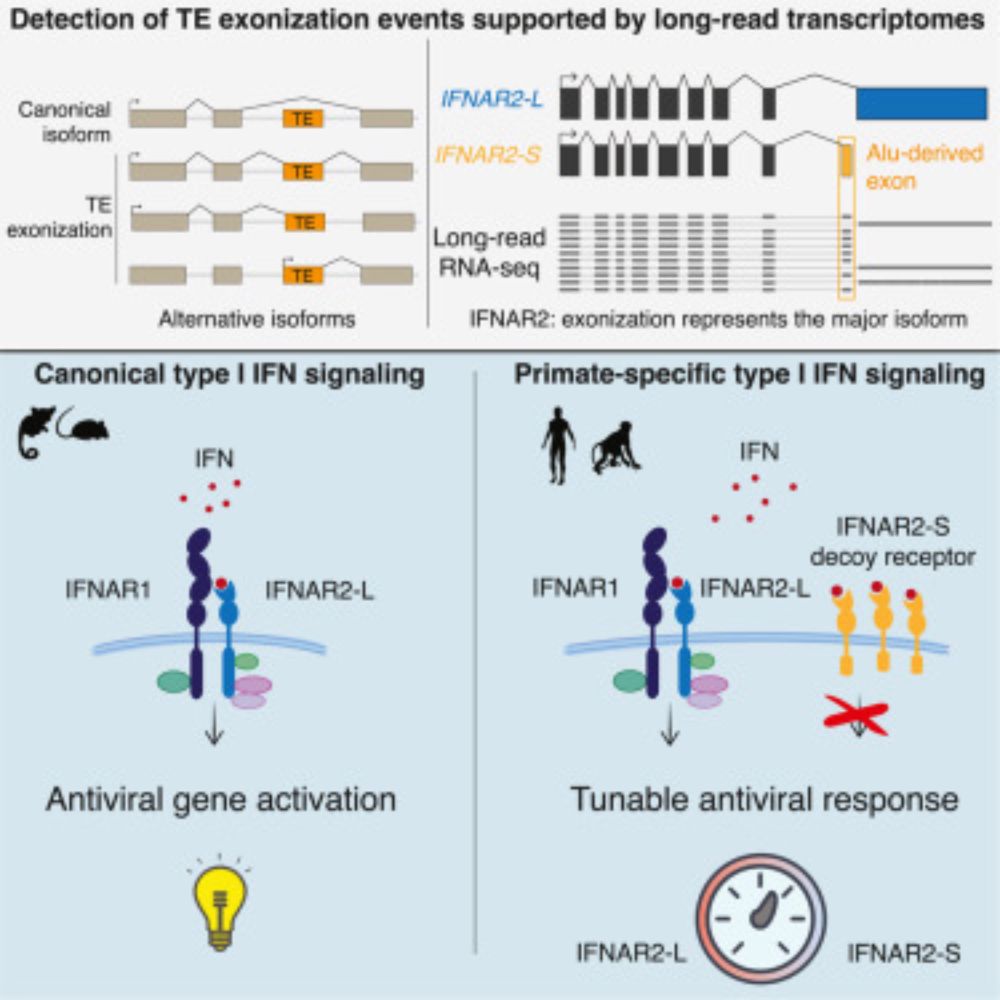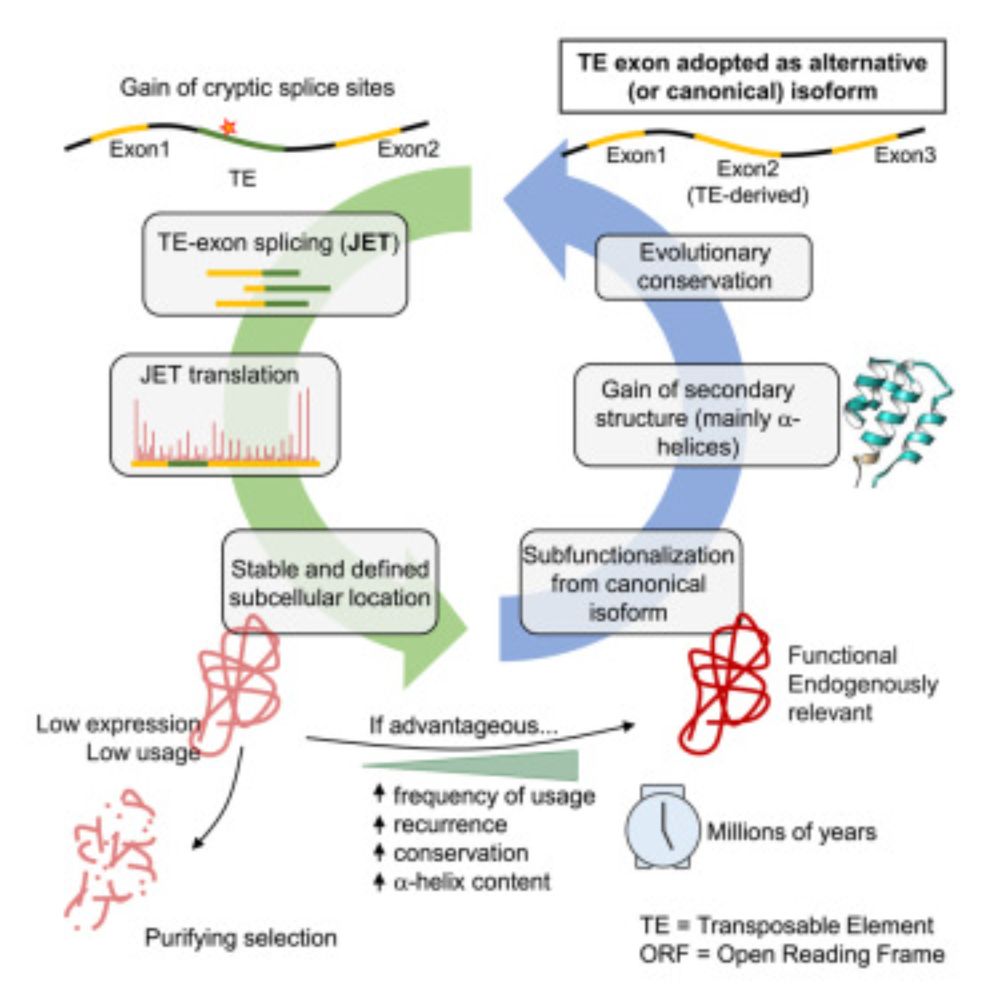
www.cell.com/cell/fulltex...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.annualreviews.org/content/jour...
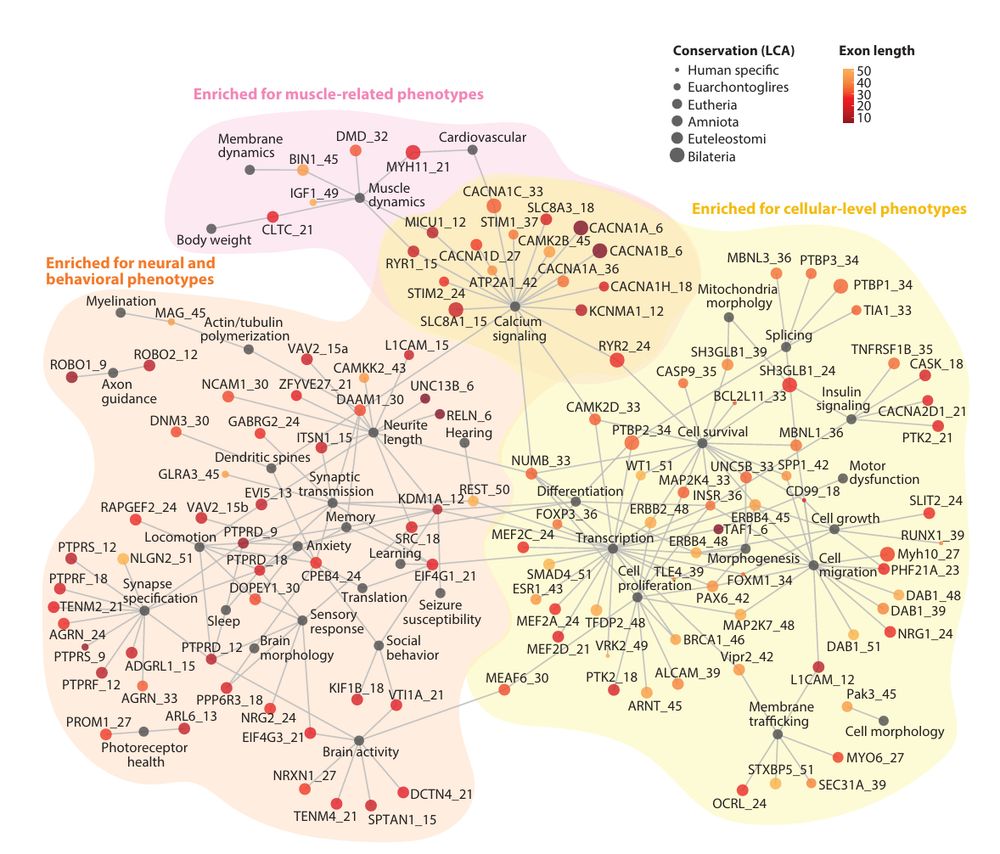
www.annualreviews.org/content/jour...
In @natmethods.nature.com we propose using "Translon" for any region decoded by the ribosome

In @natmethods.nature.com we propose using "Translon" for any region decoded by the ribosome

In our new review, @fedemantica.bsky.social and I argue we are missing the most prevalent one: specialization. And the same applies to alternative splicing! 1/7
tinyurl.com/45k7kbmp
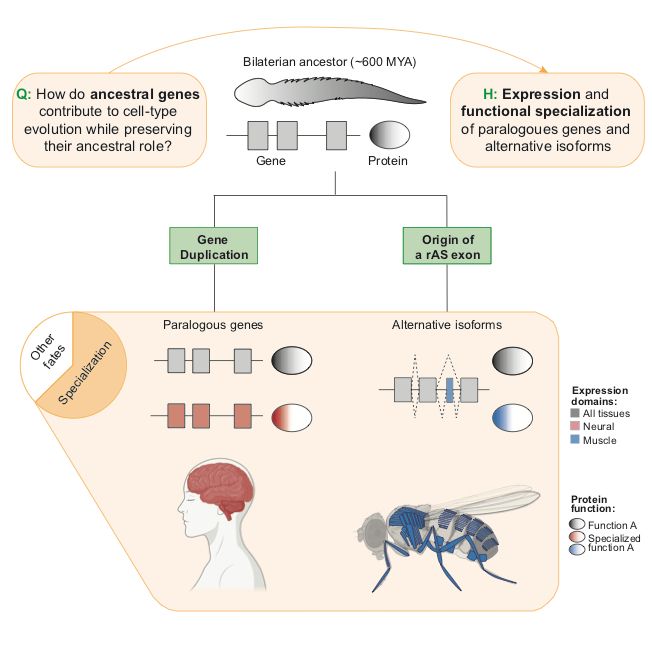
In our new review, @fedemantica.bsky.social and I argue we are missing the most prevalent one: specialization. And the same applies to alternative splicing! 1/7
tinyurl.com/45k7kbmp
More here: www.sciencedirect.com/science/arti...
#CD8 #TCellExhaustion #TNBC

More here: www.sciencedirect.com/science/arti...
#CD8 #TCellExhaustion #TNBC
Editora: @inmayruela.bsky.social
sebbm.es/revista/nume...

Editora: @inmayruela.bsky.social
sebbm.es/revista/nume...
As we wrap up our campaign, we celebrate the incredible contributions of women in science and beyond. 🌟
A big shoutout to the amazing women of the #yefis Task Force! ✨
Let’s keep inspiring and empowering each other. 💜
#WomensDay #WomenInScience #WomenInStem

As we wrap up our campaign, we celebrate the incredible contributions of women in science and beyond. 🌟
A big shoutout to the amazing women of the #yefis Task Force! ✨
Let’s keep inspiring and empowering each other. 💜
#WomensDay #WomenInScience #WomenInStem
firstwordpharma.com/story/5936238

firstwordpharma.com/story/5936238

This year’s theme: "10 Years of Discovery in Immunotherapy." We’ve put together a stellar lineup of speakers—don’t miss it!
Free registration @ bit.ly/3WFj0Hc
#ImmunoSchool #Immunotherapy #CancerResearch

This year’s theme: "10 Years of Discovery in Immunotherapy." We’ve put together a stellar lineup of speakers—don’t miss it!
Free registration @ bit.ly/3WFj0Hc
#ImmunoSchool #Immunotherapy #CancerResearch
www.cell.com/cell-reports...
www.cell.com/cell-reports...
1. You need to have someone you can talk to
2. One-on-one discussions are the best
3. Think about discussions as improvisations and use the ‘yes, and’ rule
4. Create a safe space so both of you feel open to saying seemingly silly things

1. You need to have someone you can talk to
2. One-on-one discussions are the best
3. Think about discussions as improvisations and use the ‘yes, and’ rule
4. Create a safe space so both of you feel open to saying seemingly silly things

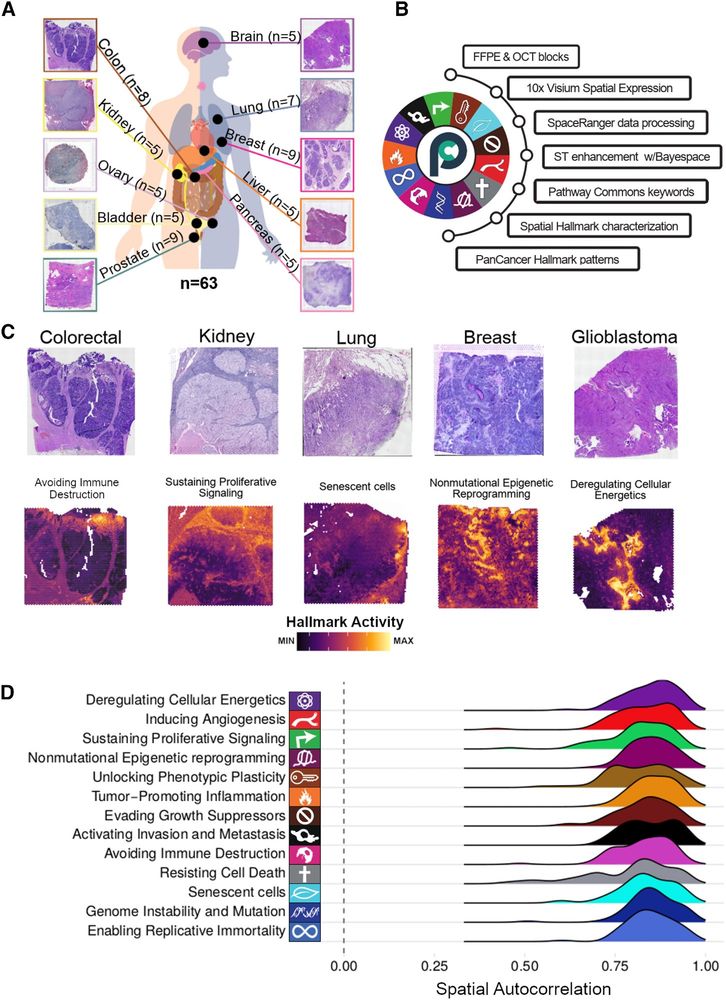
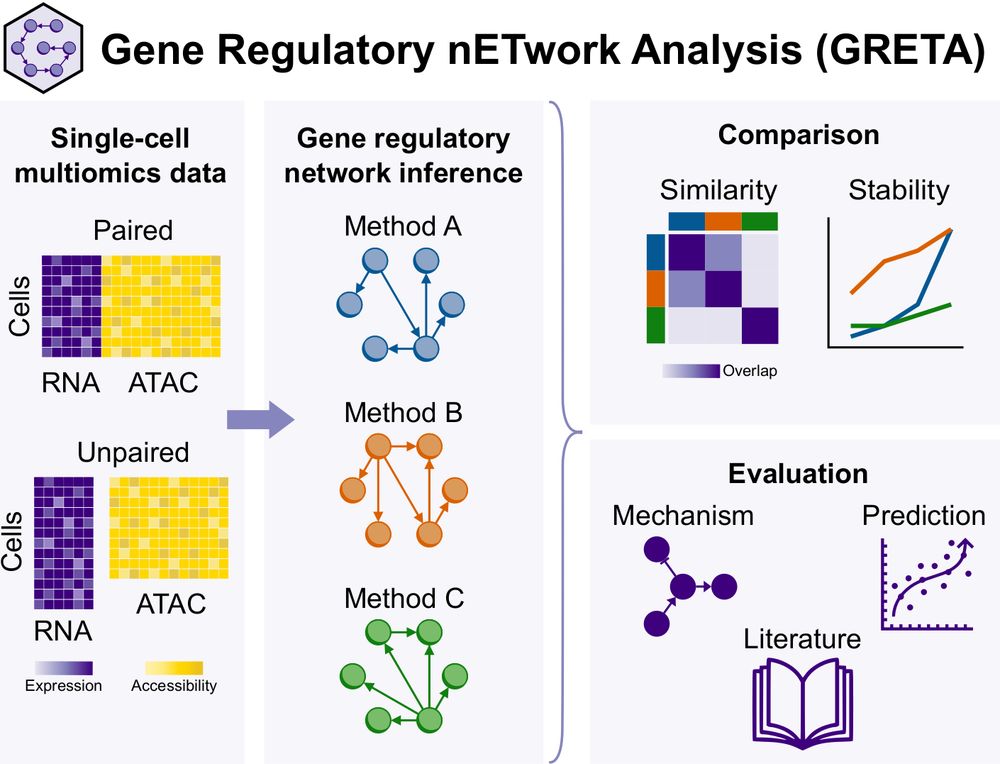
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta

Read full publication ⤵️
www.cell.com/cell/fulltext/S0092-8674%2824%2901328-X

Read full publication ⤵️
www.cell.com/cell/fulltext/S0092-8674%2824%2901328-X

📖 Descobreix tota la història de l'ACCC a accc.cat/lassociacio/#historia

📖 Descobreix tota la història de l'ACCC a accc.cat/lassociacio/#historia
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...