David A. Russo
@bluegreendr.bsky.social
80 followers
110 following
6 posts
Postdoc @ Uni Jena, cyanobacteria aficionado, passion for everything secreted #cyanobacteria #proteins #metabolites
Website: https://darusso.de/
Posts
Media
Videos
Starter Packs
Reposted by David A. Russo
Reposted by David A. Russo
David A. Russo
@bluegreendr.bsky.social
· Jul 10

Testing eukaryotic routes for heterologous production of ketocarotenoids in cyanobacteria
Abstract. Pigments and dyes are widely used in multiple industries. Ketocarotenoids are a highly sought-after group of pigments with a dark-red hue and str
academic.oup.com
Reposted by David A. Russo
Cyanotrans
@cyanotrans.bsky.social
· Jul 7

Beyond movement: the dynamic roles of Type IV pili in cyanobacterial life | Journal of Bacteriology
The advent of oxygenic photosynthesis by ancient cyanobacteria approximately 2.4 billion
years ago profoundly transformed Earth’s landscape. Today, cyanobacteria inhabit a
vast array of aquatic habita...
journals.asm.org
Reposted by David A. Russo
Wolfgang Hess
@cyanolab.bsky.social
· Jun 26
Reposted by David A. Russo
Laura T. Wey
@lauratwey.bsky.social
· Jun 3
Reposted by David A. Russo
Reposted by David A. Russo
Tomkey_kong
@tomkey-kong.bsky.social
· May 6

Gram-scale production of 4-vinyl guaiacol in the fast-growing phototrophic cyanobacterium Synechococcus sp. PCC 11901
Whole-cell catalysis in cyanobacteria allows the transformation of light energy into chemical energy by co-factor recycling and in situ production of oxygen by photosynthesis, requiring only light, CO...
dx.doi.org
Reposted by David A. Russo
Kedrov Lab @ HHU
@kedrov-lab.bsky.social
· May 27

Substrate-induced assembly and functional mechanism of the bacterial membrane protein insertase SecYEG-YidC
The universally conserved Sec translocon and the YidC/Oxa1-type insertases mediate biogenesis of alpha-helical membrane proteins, but the molecular basis of their cooperation has remained disputed over decades. A recent discovery of a multi-subunit insertase in eukaryotes has raised the question about the architecture of the putative bacterial ortholog SecYEG-YidC and its functional mechanism. Here, we combine cryogenic electron microscopy with cell-free protein synthesis in nanodiscs to visualize biogenesis of the polytopic membrane protein NuoK, the subunit K of NADH-quinone oxidoreductase, that requires both SecYEG and YidC for insertion. We demonstrate that YidC is recruited to the back of the translocon at the late stage of the substrate insertion, in resemblance to the eukaryotic system, and in vivo experiments indicate that the complex assembly is vital for the cells. The nascent chain does not utilize the lateral gate of SecYEG, but enters the lipid membrane at the SecYE-YidC interface, with YidC being the primary insertase. SecYEG-YidC complex promotes folding of the nascent helices at the interface prior their insertion, so the examined cellular pathway follows the fundamental thermodynamic principles of membrane protein folding. Our data provide the first detailed insight on the elusive insertase machinery in the physiologically relevant environment, highlight the importance of the nascent chain for its assembly, and prove the evolutionary conservation of the gate-independent insertion route. ### Competing Interest Statement The authors have declared no competing interest. Deutsche Forschungsgemeinschaft, https://ror.org/018mejw64, Ke1879/3, 267205415 (CRC 1208) European Research Council, https://ror.org/0472cxd90, CRYOTRANSLATION
www.biorxiv.org
Reposted by David A. Russo
Reposted by David A. Russo
David A. Russo
@bluegreendr.bsky.social
· Mar 11
Elke Dittmann
@dittmannelke.bsky.social
· Mar 10
Competition and interdependence define interactions of Nostoc sp. and Agrobacterium sp. under inorganic carbon limitation
npj Biofilms and Microbiomes - Competition and interdependence define interactions of Nostoc sp. and Agrobacterium sp. under inorganic carbon limitation
rdcu.be
Zedler Lab
@synbiojazz.bsky.social
· Mar 6
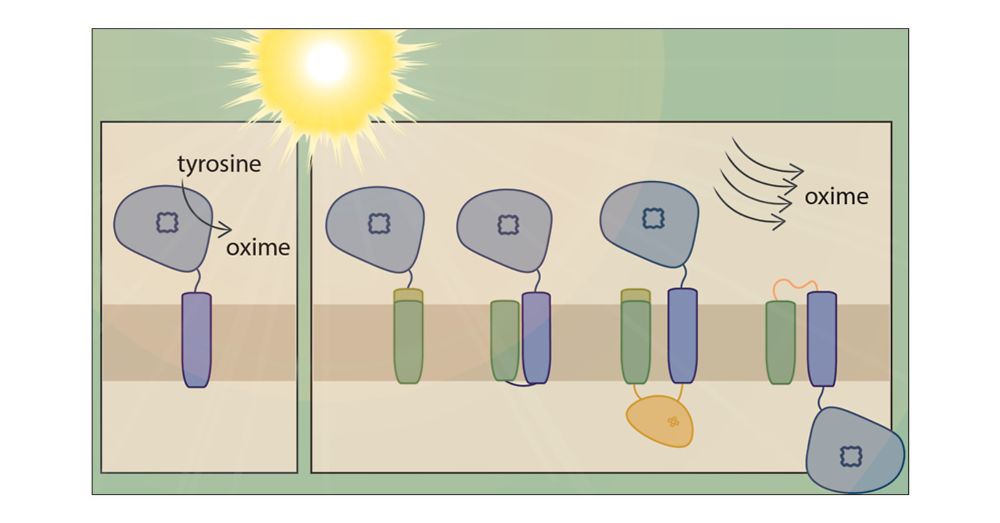
Thylakoid Targeting Improves Stability of a Cytochrome P450 in the Cyanobacterium Synechocystis sp. PCC 6803
Plants produce a large array of natural products of biotechnological interest. In many cases, these compounds are naturally produced at low titers and involve complex biosynthetic pathways, which often include cytochrome P450 enzymes. P450s are known to be difficult to express in traditional heterotrophic chassis. However, cyanobacteria have shown promise as a sustainable alternative for the heterologous expression of P450s and light-driven product biosynthesis. In this study, we explore strategies for improving plant P450 stability and membrane insertion in cyanobacteria. The widely used model cyanobacterium Synechocystis sp. PCC 6803 was chosen as the host, and the well-studied P450 CYP79A1 from the dhurrin pathway of Sorghum bicolor was chosen as the model enzyme. Combinations of the P450 fused with individual elements (e.g., signal peptide, transmembrane domain) or the full length cyanobacterial, thylakoid-localized, protein PetC1 were designed. All generated CYP79A1 variants led to oxime production. Our data show that strains producing CYP79A1 variants with elements of PetC1 improved thylakoid targeting. In addition, chlorophyll-normalized oxime levels increased, on average, up to 18 times compared to the unmodified CYP79A1. These findings offer promising strategies to improve heterologous P450 expression in cyanobacteria and can ultimately contribute to advancing light-driven biocatalysis in cyanobacterial chassis.
pubs.acs.org
Reposted by David A. Russo