Charlie Hale
@charleshale.bsky.social
230 followers
770 following
16 posts
PhD student in the Buckler Lab at Cornell. Evolution, genomics, climate change, agriculture.
Posts
Media
Videos
Starter Packs
Pinned
Charlie Hale
@charleshale.bsky.social
· Apr 27

Extensive modulation of a conserved cis-regulatory code across 589 grass species
The growing availability of genomes from non-model organisms offers new opportunities to identify functional loci underlying trait variation through comparative genomics. While cis-regulatory regions ...
www.biorxiv.org
Reposted by Charlie Hale
Reposted by Charlie Hale
Reposted by Charlie Hale
Chenxin Li, PhD
@chenxinli2.bsky.social
· Jul 21
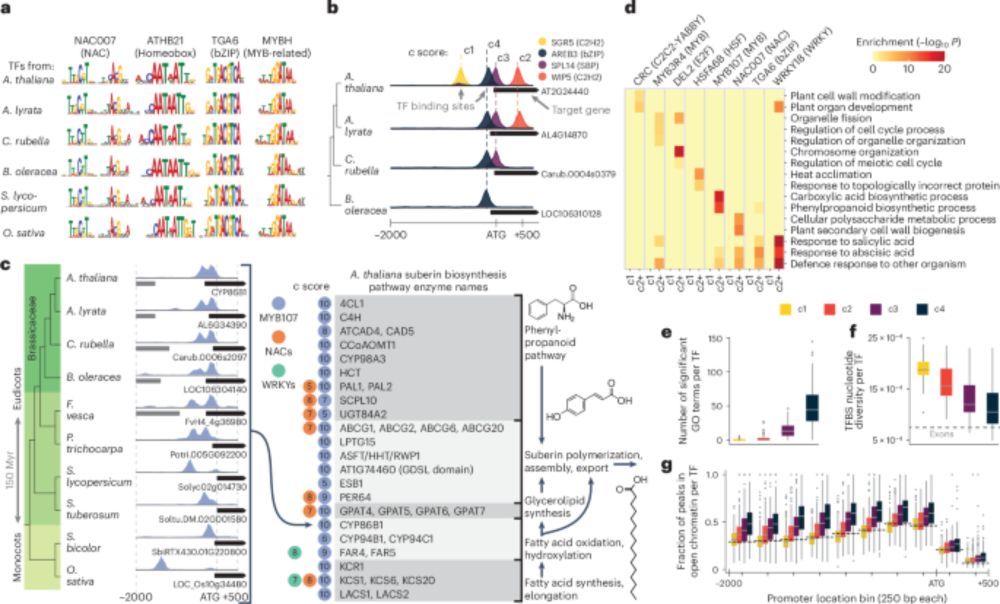
Recruitment, rewiring and deep conservation in flowering plant gene regulation - Nature Plants
A highly scalable approach is used to generate 3,000 genome-wide maps of transcription factor binding in ten flowering plants, along with multi-species single-nucleus RNA-seq atlases. Together, the re...
www.nature.com
Reposted by Charlie Hale
Reposted by Charlie Hale
Erin Morrow
@erinmorrow.bsky.social
· Jun 16
Reposted by Charlie Hale
Reposted by Charlie Hale
Reposted by Charlie Hale
Marc Dionne
@marcsdionne.bsky.social
· Jun 3
Drosophila Genetic Database
The Drosophila Genetic Database, FlyBase, is on the brink of collapse due to the sudden termination of the FlyBase NIH grant, which includes salaries for 5 literature curators based at the University ...
www.philanthropy.cam.ac.uk
Reposted by Charlie Hale
Charlie Hale
@charleshale.bsky.social
· May 26

Grass Rhizome Proteomics Reveals Convergent Freezing-Tolerance Strategies
Early maize planting requires cold tolerance in temperate regions, which elite maize lacks. Extending the growing season could boost productivity along with more efficient nutrient use. Wild PACMAD gr...
www.biorxiv.org
Reposted by Charlie Hale
Reposted by Charlie Hale
Reposted by Charlie Hale
Reposted by Charlie Hale
Charlie Hale
@charleshale.bsky.social
· Apr 27
Charlie Hale
@charleshale.bsky.social
· Apr 27

Extensive modulation of a conserved cis-regulatory code across 589 grass species
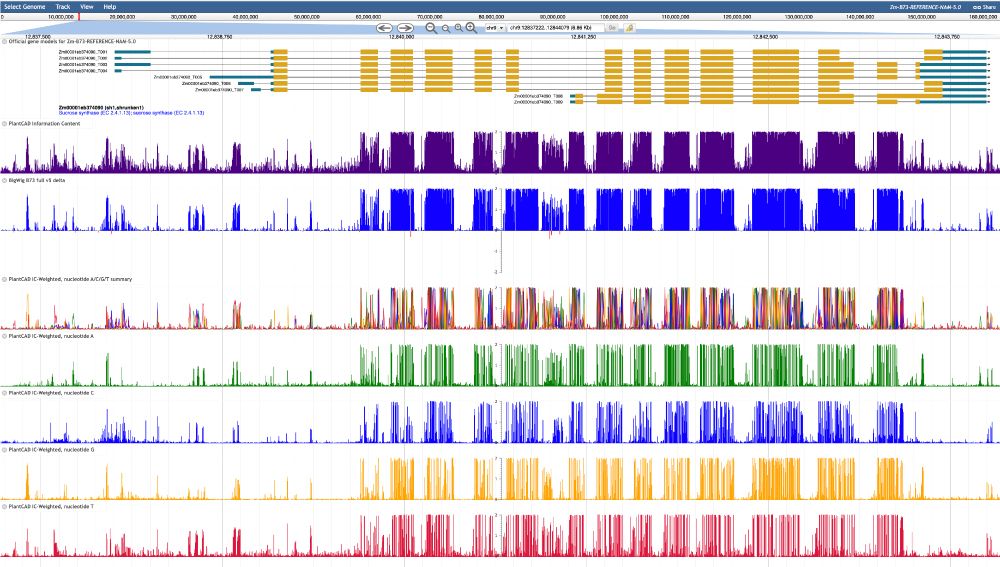
The growing availability of genomes from non-model organisms offers new opportunities to identify functional loci underlying trait variation through comparative genomics. While cis-regulatory regions drive much of phenotypic evolution, linking them to specific functions remains challenging. We identified 514 cis-regulatory motifs enriched in regulatory regions of five diverse grass species, with 73% consistently enriched across all, suggesting a deeply conserved regulatory code. We then quantified conservation of specific motif instances across 589 grass species, revealing widespread gain and loss over evolutionary time. Conservation declined rapidly over the first few million years of divergence, yet ~50% of motif instances were conserved back to the origin of grasses ~100 million years ago. Conservation patterns varied by gene class, with modestly higher conservation at transcription factor genes. To test for adaptive cis-regulatory changes, we used phylogenetic mixed models to identify motif gains and losses associated with ecological niche transitions. Our models revealed polygenic adaptation across 810 motif-orthogroup combinations, including convergent gains of HSF/GARP motifs at an Alpha-N-acetylglucosaminidase gene associated with adaptation to temperate environments. Our results support a model in which cis-regulatory evolution involves extensive turnover of individual binding site instances while largely preserving transcription factors' binding preferences. Convergent cis-regulatory changes at hundreds to thousands of genes likely contribute to environmental adaptation. Our results highlight the potential of comparative genomics and phylogenetic mixed models to reveal the genetic basis of complex traits. ### Competing Interest Statement The authors have declared no competing interest. National Science Foundation, https://ror.org/021nxhr62, 1822330, 2139899, 1907343 United States Department of Agriculture, , 8062-21000-052-004-A, 8062-21000-052-000-D National Institutes of Health, , 1R00GM144742
www.biorxiv.org
Charlie Hale
@charleshale.bsky.social
· Apr 27
Charlie Hale
@charleshale.bsky.social
· Apr 27
Charlie Hale
@charleshale.bsky.social
· Apr 27

Extensive modulation of a conserved cis-regulatory code across 589 grass species
The growing availability of genomes from non-model organisms offers new opportunities to identify functional loci underlying trait variation through comparative genomics. While cis-regulatory regions ...
www.biorxiv.org