David Ochoa
@d0choa.bsky.social
310 followers
350 following
22 posts
Target discovery | Computational Biology | Human Disease Genetics | ML | Cloud computing - @opentargets.org
- @ebi.embl.org
Posts
Media
Videos
Starter Packs
Reposted by David Ochoa
Konrad
@konradjk.bsky.social
· 20d

Pan-UK Biobank genome-wide association analyses enhance discovery and resolution of ancestry-enriched effects - Nature Genetics
Genome-wide analyses for 7,266 traits leveraging data from several genetic ancestry groups in UK Biobank identify new associations and enhance resources for interpreting risk variants across diverse p...
www.nature.com
Reposted by David Ochoa
David Ochoa
@d0choa.bsky.social
· Jun 18

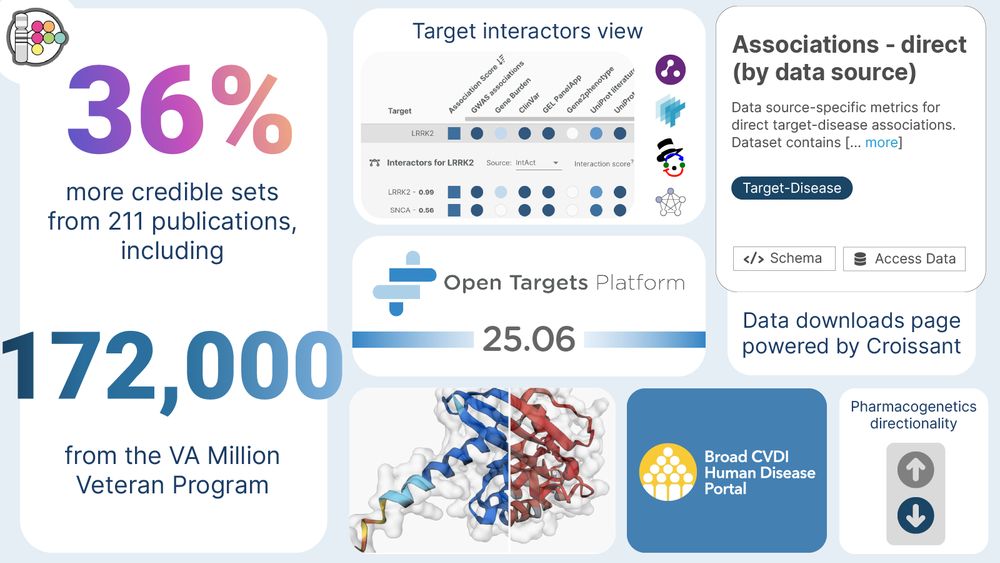
25.06 Platform Release Q&A | LinkedIn
Join our Q&A session about the Open Targets Platform June release.
The team will run through the main points of the release, after which they will open the floor to questions.
The walkthrough will b...
www.linkedin.com
David Ochoa
@d0choa.bsky.social
· Jun 18
David Ochoa
@d0choa.bsky.social
· Jun 18
David Ochoa
@d0choa.bsky.social
· Jun 18
David Ochoa
@d0choa.bsky.social
· Jun 18
David Ochoa
@d0choa.bsky.social
· Jun 18
Reposted by David Ochoa
Reposted by David Ochoa
Reposted by David Ochoa
David Ochoa
@d0choa.bsky.social
· Mar 20
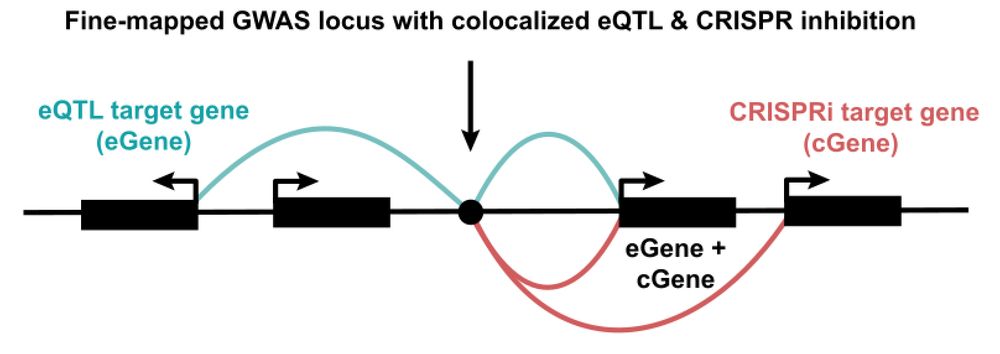
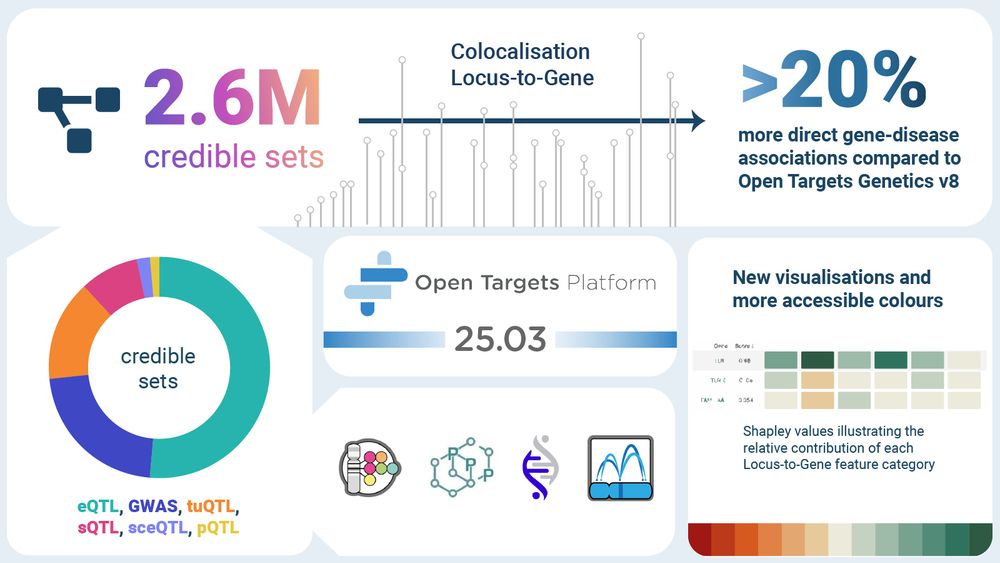
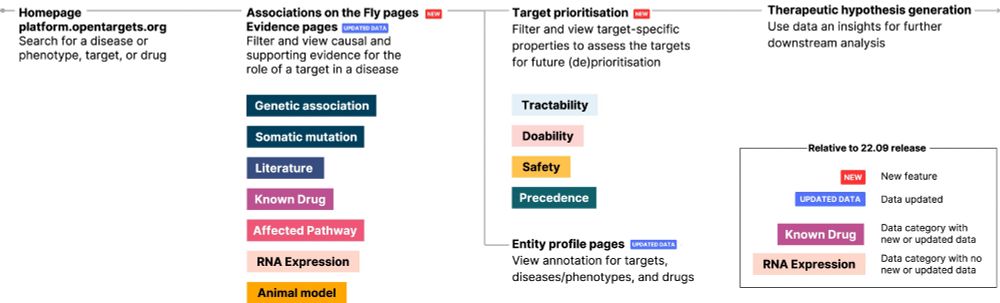
A step-change in common disease genetics in the Open Targets Platform
We identified an opportunity to create a unified resource to seamlessly access human genetic and target discovery information. The Platform now integrates and evaluates gene-disease associations from ...
blog.opentargets.org
David Ochoa
@d0choa.bsky.social
· Dec 17
Reposted by David Ochoa
Jeff Spence
@jeffspence.github.io
· Dec 17

Specificity, length, and luck: How genes are prioritized by rare and common variant association studies
Standard genome-wide association studies (GWAS) and rare variant burden tests are essential tools for identifying trait-relevant genes. Although these methods are conceptually similar, we show by anal...
www.biorxiv.org
Reposted by David Ochoa
Reposted by David Ochoa