Robert Hänsel-Hertsch
@epistrucstab.bsky.social
110 followers
180 following
21 posts
Junior Group Leader, Center for Molecular Medicine Cologne, University of Cologne, DE
Posts
Media
Videos
Starter Packs
Reposted by Robert Hänsel-Hertsch
Reposted by Robert Hänsel-Hertsch
Waggoner Lab
@labwaggoner.bsky.social
· Jul 29
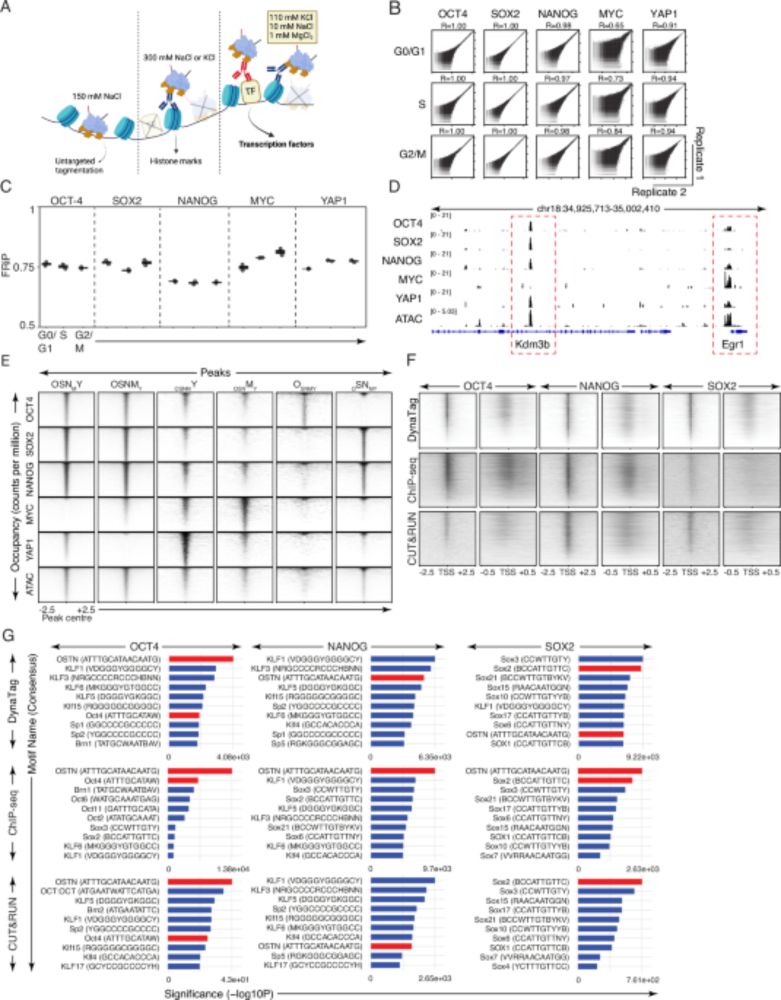
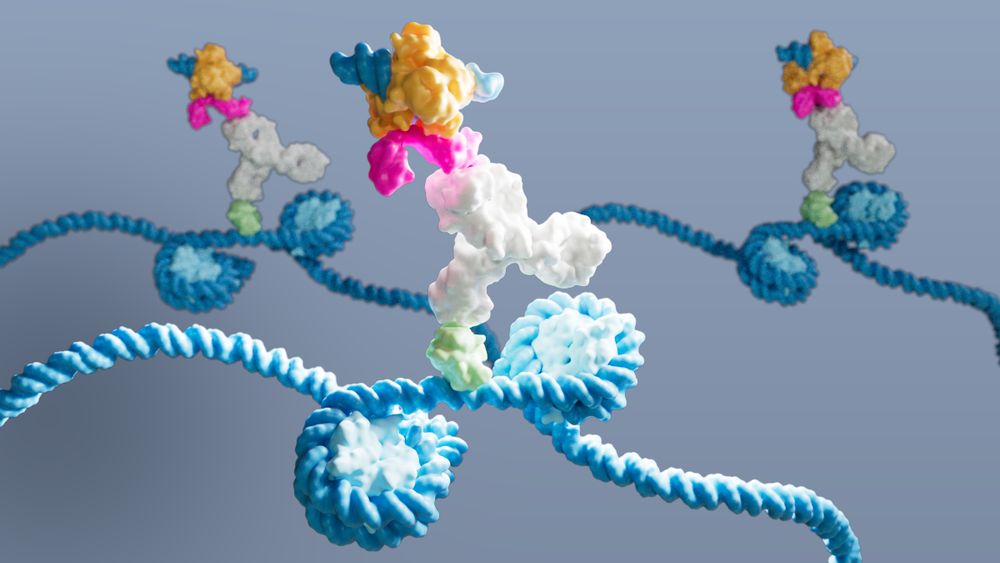
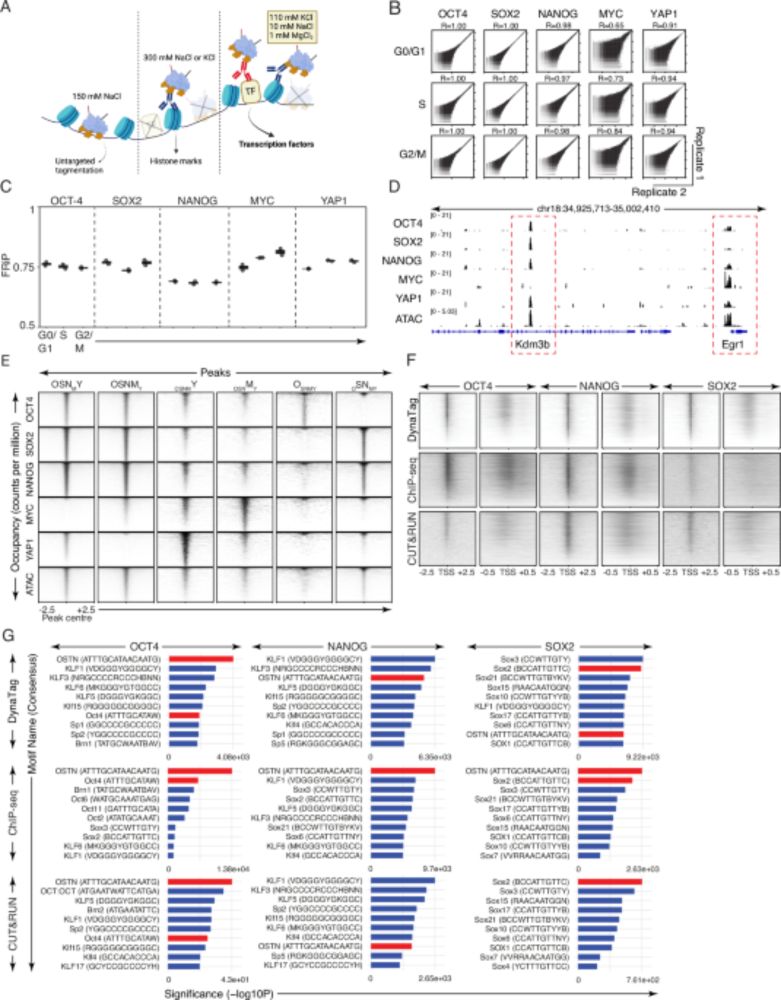
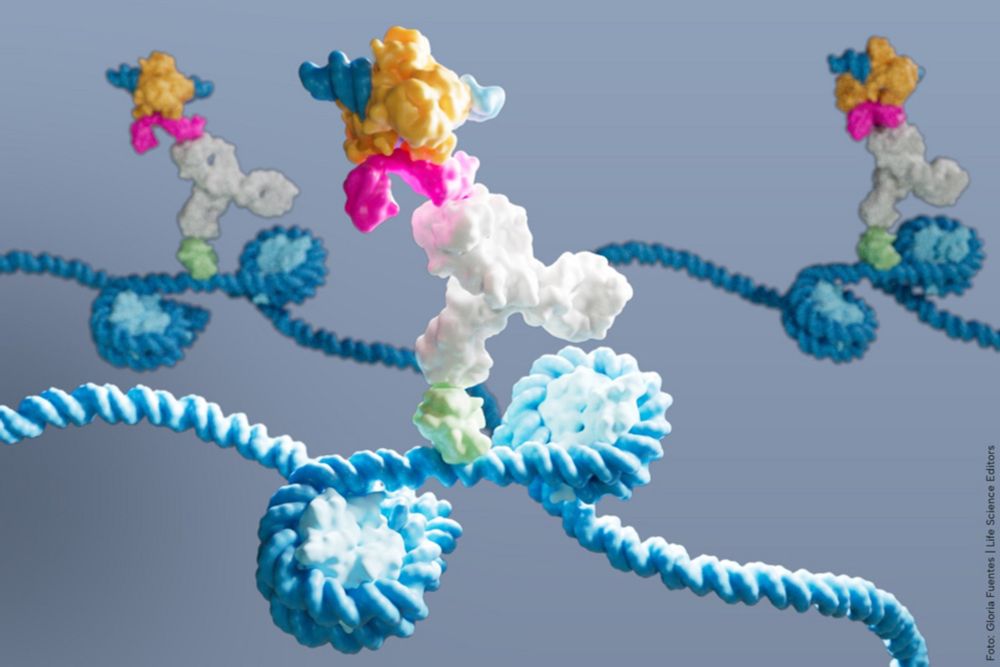
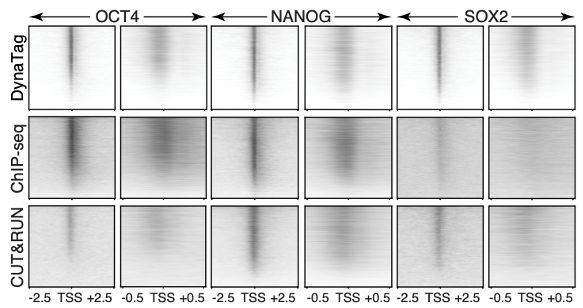
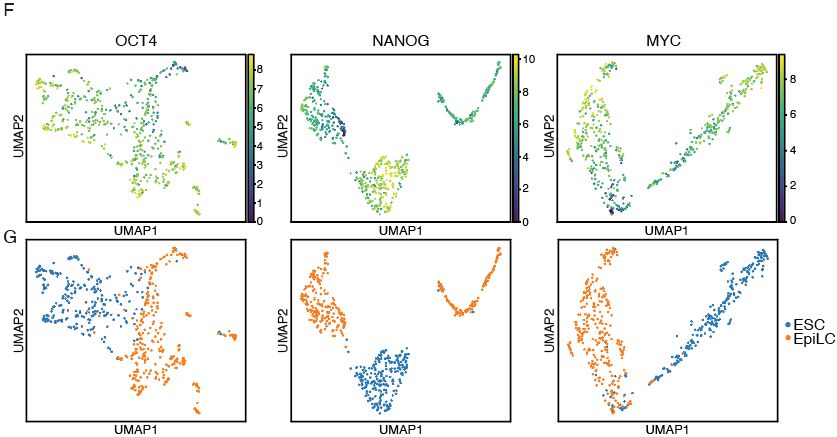
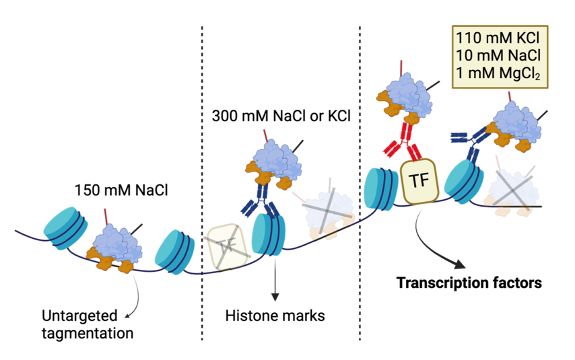
DynaTag for efficient mapping of transcription factors in low-input samples and at single-cell resolution - Nature Communications
Transcription factors shape cell identity, but mapping their genomic targets remains challenging. Here the authors present DynaTag, a modified CUT&Tag method for profiling TF occupancy in bulk and...
www.nature.com
Reposted by Robert Hänsel-Hertsch
Reposted by Robert Hänsel-Hertsch
Reposted by Robert Hänsel-Hertsch
CRC1678
@crc1678.bsky.social
· Jul 28
Reposted by Robert Hänsel-Hertsch
Sonhita 🌞
@sonhita.bsky.social
· Jul 28
Reposted by Robert Hänsel-Hertsch
Reposted by Robert Hänsel-Hertsch