Christof Gebhardt
@gebhardtlab.bsky.social
140 followers
210 following
18 posts
Physics at Ulm University | live-cell/organism single-molecule microscopy | super-resolution microscopy | gene regulation | chromatin architecture | embryo development
Posts
Media
Videos
Starter Packs
Reposted by Christof Gebhardt
Reposted by Christof Gebhardt
Di Jiang
@dijiang319.bsky.social
· May 29
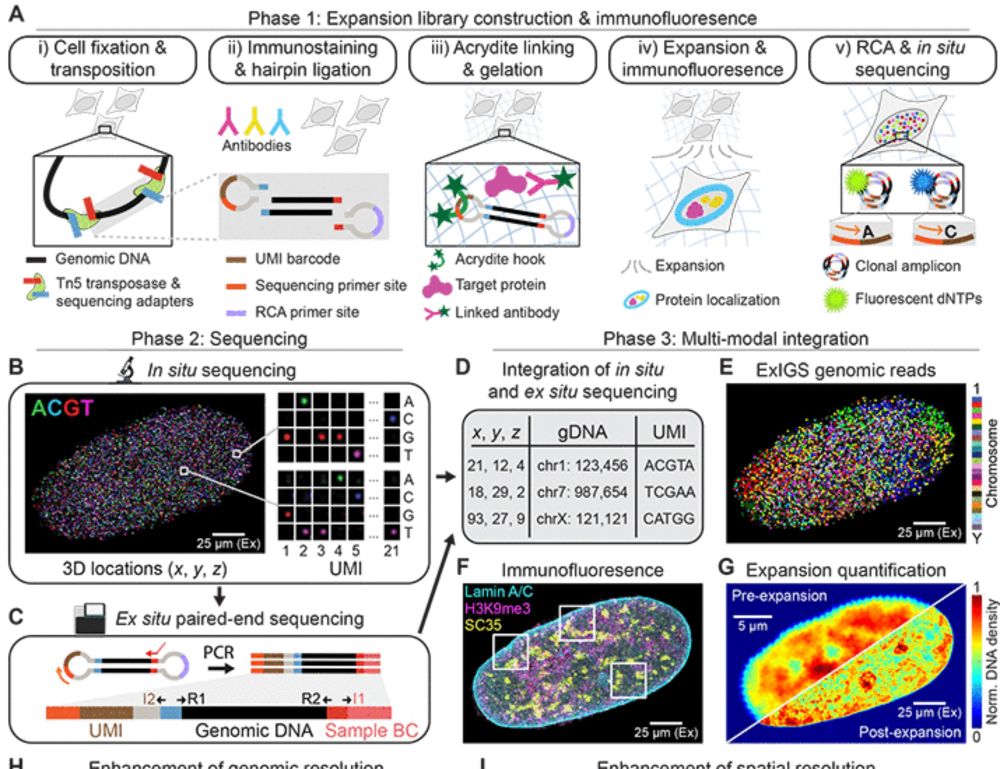
Expansion in situ genome sequencing links nuclear abnormalities to aberrant chromatin regulation
Microscopy and genomics are used to characterize cell function, but approaches to connect the two types of information are lacking, particularly at subnuclear resolution. Here, we describe expansion i...
www.science.org
Reposted by Christof Gebhardt
Reposted by Christof Gebhardt
Reposted by Christof Gebhardt
David Suter
@davidsuter.bsky.social
· Mar 7

Core passive and facultative mTOR-mediated mechanisms coordinate mammalian protein synthesis and decay
The maintenance of cellular homeostasis requires tight regulation of proteome concentration and composition. To achieve this, protein production and elimination must be robustly coordinated. However, ...
www.biorxiv.org
Reposted by Christof Gebhardt
Reposted by Christof Gebhardt
Reposted by Christof Gebhardt
Reposted by Christof Gebhardt
Reposted by Christof Gebhardt
David Suter
@davidsuter.bsky.social
· Feb 13
Reposted by Christof Gebhardt