Jerónimo Rodríguez-Beltrán
@jerorb.bsky.social
920 followers
570 following
65 posts
We study Antibiotic Resistance Ecology and Evolution · Ramón y Cajal University Hospital · Madrid · www.evodynamicslab.com
Posts
Media
Videos
Starter Packs
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Eduardo Rocha
@epcrocha.bsky.social
· Sep 8
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
DaganLab
@daganlab.bsky.social
· Sep 5

W 3 professorship in Molecular and Evolutionary Zoology - Kiel, Schleswig-Holstein (DE) job with Christian-Albrechts-Universität zu Kiel | 12844416
Kiel University aims to attract more qualified women to professorship positions. The Institute of Zoology at the Faculty of Mathematics and Natural...
www.nature.com
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Michael Baym
@baym.lol
· Sep 1

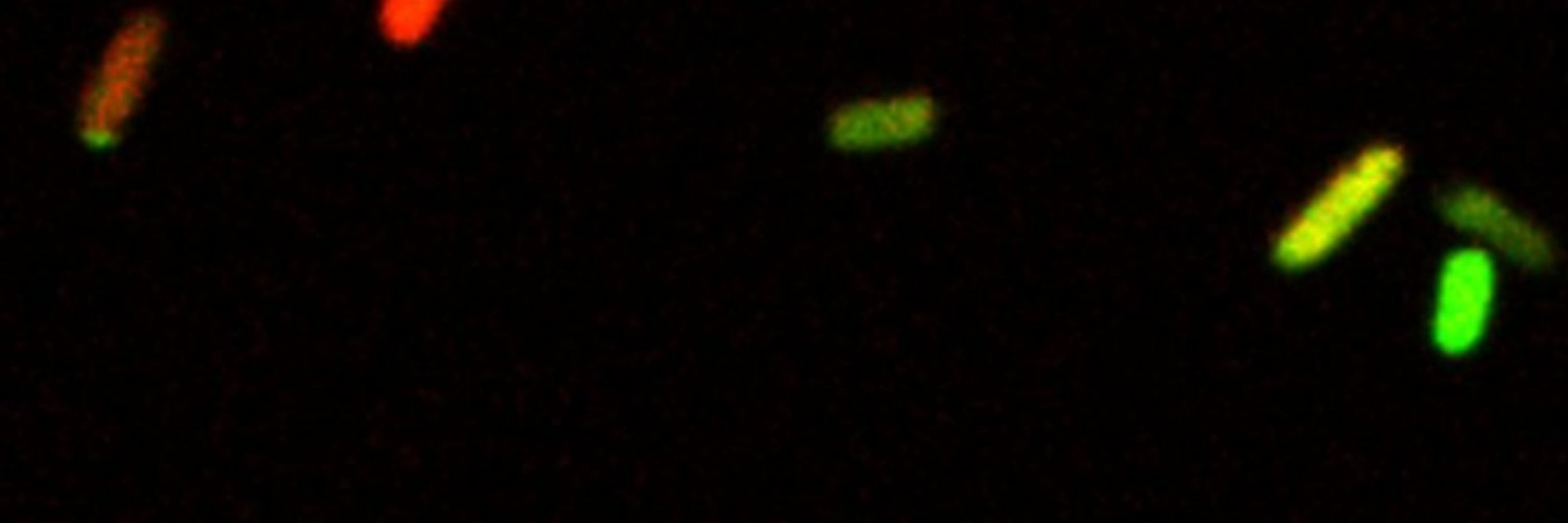
Genomic resistance in historical clinical isolates increased in frequency and mobility after the age of antibiotics
Antibiotic resistance is frequently observed shortly after the clinical introduction of an antibiotic. Whether and how frequently that resistance occurred before the introduction is harder to determin...
www.microbiologyresearch.org
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
James McInerney
@jomcinerney.bsky.social
· Aug 29

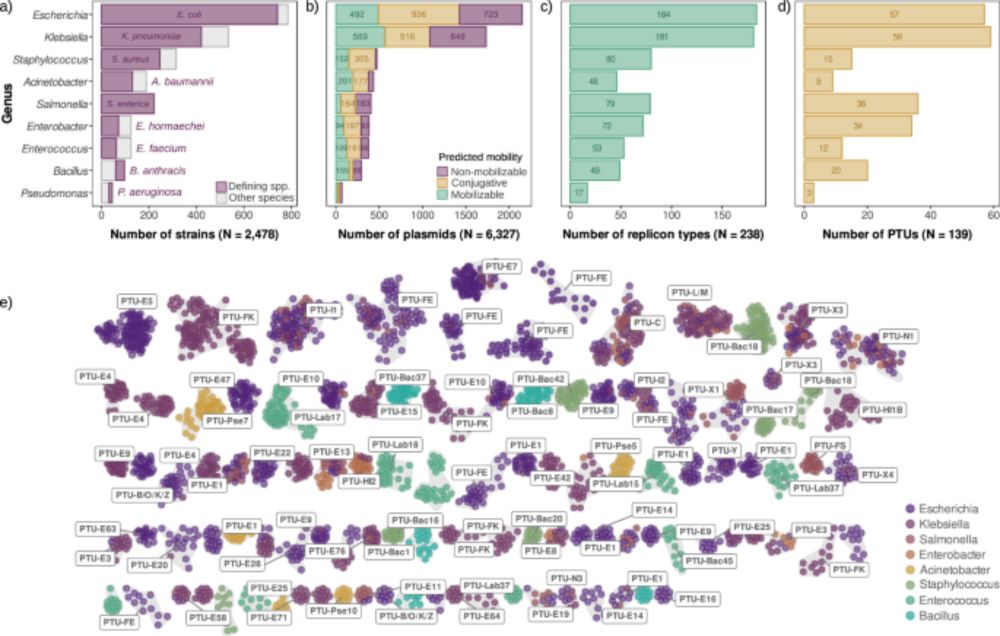
Genomic constraints shape the evolution of alternative routes to drug resistance in prokaryotes
Background Variation within the prokaryotic pangenome is not random, and natural selection that favours particular combinations of genes appears to dominate over random drift. What is less clear is wh...
www.biorxiv.org
Reposted by Jerónimo Rodríguez-Beltrán
Reposted by Jerónimo Rodríguez-Beltrán
Eduardo Rocha
@epcrocha.bsky.social
· Aug 27
Reposted by Jerónimo Rodríguez-Beltrán