Posts
Media
Videos
Starter Packs
Reposted by Robert Kofler
Reposted by Robert Kofler
Matt Beaumont
@mbeaum.bsky.social
· Aug 28

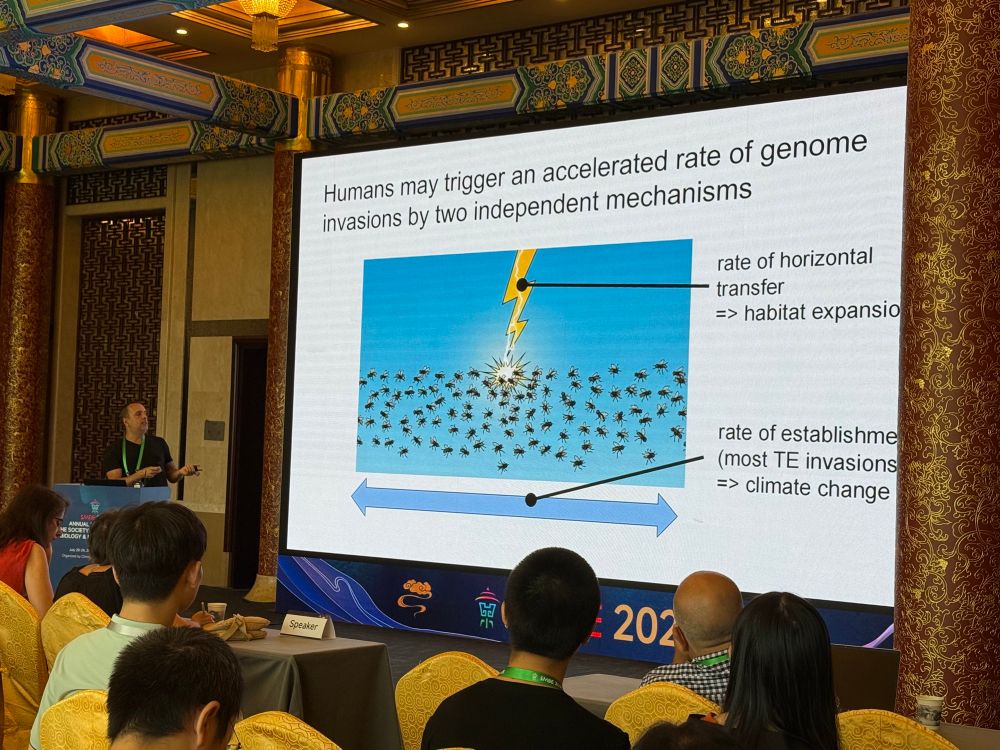
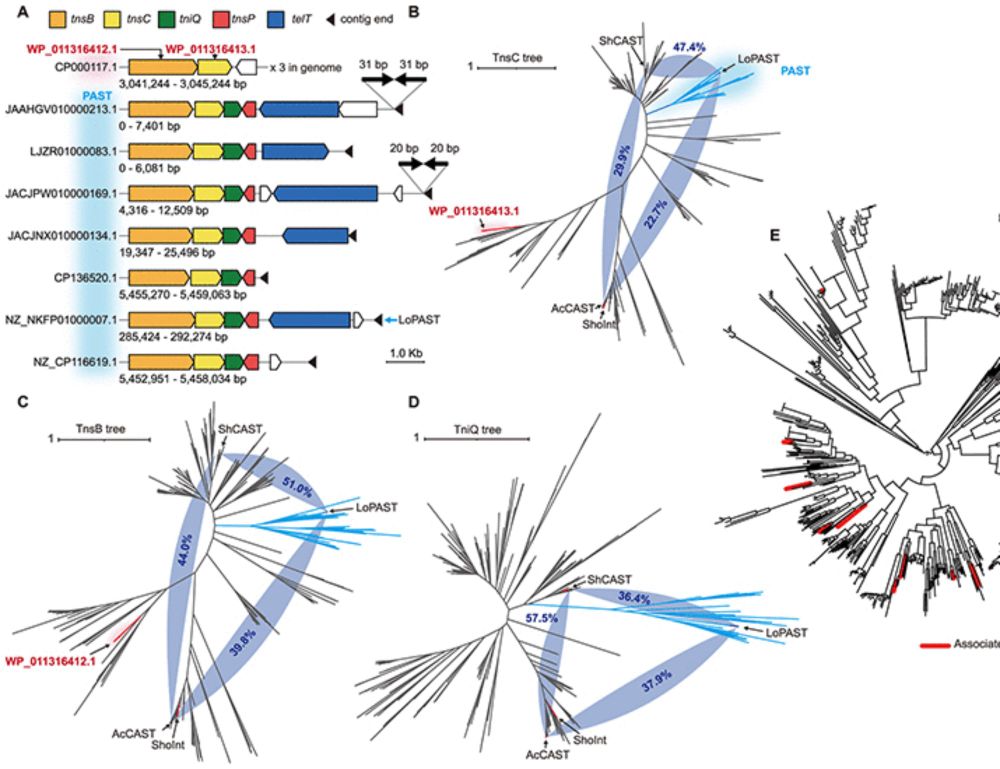
Rapid emergence of non-autonomous elements may stop P-element invasions in the absence of a piRNA-based host defence
Author summary Transposable elements (TEs) are short, self-replicating DNA sequences found in nearly all genomes. While they can be harmful to their hosts, many organisms have evolved defence systems,...
journals.plos.org
Reposted by Robert Kofler
Reposted by Robert Kofler
Reposted by Robert Kofler
Reposted by Robert Kofler
Robert Kofler
@rokofler.bsky.social
· Jun 11
Robert Kofler
@rokofler.bsky.social
· Jun 11

Spatio-temporal tracking of three novel transposable element invasions in Drosophila melanogaster over the last 30 years
Abstract. Transposable elements (TEs) are repetitive sequences capable of mobilizing within genomes, exerting a sigfinificant influence on evolution throug
academic.oup.com
Reposted by Robert Kofler
Robert Kofler
@rokofler.bsky.social
· Jun 3
Reposted by Robert Kofler
Reposted by Robert Kofler
Reposted by Robert Kofler
Reposted by Robert Kofler
Oded Rechavi
@odedrechavi.bsky.social
· Mar 7