Thom Booth
@thombooth.bsky.social
470 followers
180 following
100 posts
NNF Postdoctoral Fellow at DTU Biosustain. Interested in the discovery and evolution of biosynthetic pathways.
Posts
Media
Videos
Starter Packs
Pinned
Thom Booth
@thombooth.bsky.social
· Jul 9
Evidence supporting the first secondary chromosome in actinobacteria as a hallmark of the Embleya genus
Embleya is a genus within the family Streptomycetaceae, a group of actinobacteria with outstanding capacity for production of specialised metabolites and a strikingly complex life cycle. In this work, we sequenced the complete genome of the new species Embleya australiensis MST-11070 and validated the assembly using optical mapping. The genome of E. australiensis MST-11070 consists of a 7.1 Mb linear chromosome and three additional replicons, including a 4.2 Mb linear replicon, EEC1, significantly larger than all previously described secondary replicons from bacteria. EEC1 is typified by its similar composition to the chromosome in terms of GC-content, codon usage and gene functions. It also carries terminal inverted repeats identical to the chromosome. EEC1 is enriched in biosynthetic gene clusters (BGCs), including the only copy of the BGCs for the spore pigment and the surfactant peptide SapB, metabolites essential for the organism's lifecycle. EEC1 contains an origin of replication with at least some chromosomal properties, and its replication is likely to depend on functions provided by chromosomally located genes. Further comparison of Embleya spp. genomes suggests that EEC1-like replicons are conserved across the genus, in contrast to other known large linear extrachromosomal replicons (megaplasmids) in the order. EEC1 is thus a hallmark of the Embleya genus and is central to its evolution within the Streptomycetaceae family. We propose EEC1 as a secondary chromosome, distinct from previously described secondary chromosomes that utilise plasmid-like replication mechanisms (chromids) and the largest secondary replicon reported in bacteria, to date. ### Competing Interest Statement Ernest Lacey is a Founder, Board Member, and the Managing Director of Microbial Screening Technology Pty. Ltd. The authors declare no competing financial interests. Biotechnology and Biological Sciences Research Council, https://ror.org/00cwqg982, BB/P021506/1, BBS/E/J/000PR9790, BB/X01097X/1, BB/M011216/1 Novo Nordisk Foundation, https://ror.org/04txyc737, NNF22OC0078997
www.biorxiv.org
Reposted by Thom Booth
Thom Booth
@thombooth.bsky.social
· Jul 27
Reposted by Thom Booth
Reposted by Thom Booth
Team Thomma
@teamthomma.bsky.social
· Jul 24
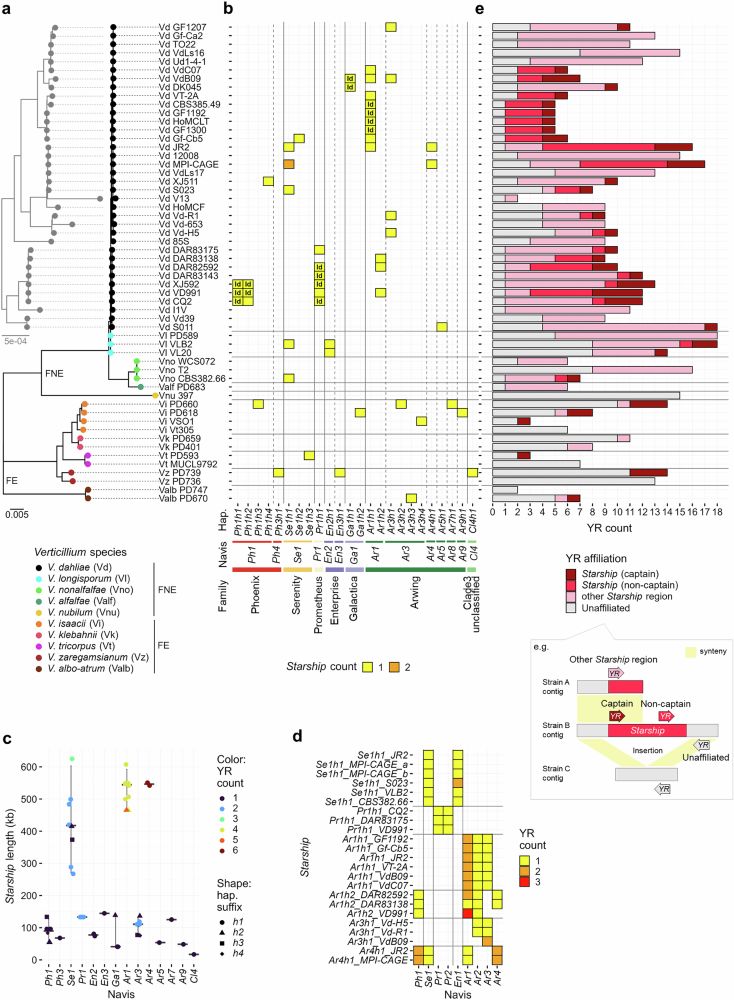
Starship giant transposons dominate plastic genomic regions in a fungal plant pathogen and drive virulence evolution
Nature Communications - Giant transposons, known as ‘Starships’, mediate horizontal gene transfer between fungal genomes. Here, Sato et al. show that Starships occupy genome regions...
rdcu.be
Reposted by Thom Booth
Keith Trujillo
@neurochicano.bsky.social
· Jul 17
Reposted by Thom Booth
Barrie Wilkinson
@barriewilks.bsky.social
· Jul 15
Reposted by Thom Booth
Susan Schlimpert
@s-lab.bsky.social
· Jul 10

Independent Fellowship in Plant-Associated Microbial Interactions | John Innes Centre
An exciting opportunity for an Independent Fellowship in Plant-Associated Microbial Interactions has arisen at the John Innes Centre. To read the full job description for this role…
www.jic.ac.uk
Reposted by Thom Booth
Tung Le
@tunglejic.bsky.social
· Jul 10
Susan Schlimpert
@s-lab.bsky.social
· Jul 10

Independent Fellowship in Plant-Associated Microbial Interactions | John Innes Centre
An exciting opportunity for an Independent Fellowship in Plant-Associated Microbial Interactions has arisen at the John Innes Centre. To read the full job description for this role…
www.jic.ac.uk
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Thom Booth
@thombooth.bsky.social
· Jul 9
Evidence supporting the first secondary chromosome in actinobacteria as a hallmark of the Embleya genus
Embleya is a genus within the family Streptomycetaceae, a group of actinobacteria with outstanding capacity for production of specialised metabolites and a strikingly complex life cycle. In this work, we sequenced the complete genome of the new species Embleya australiensis MST-11070 and validated the assembly using optical mapping. The genome of E. australiensis MST-11070 consists of a 7.1 Mb linear chromosome and three additional replicons, including a 4.2 Mb linear replicon, EEC1, significantly larger than all previously described secondary replicons from bacteria. EEC1 is typified by its similar composition to the chromosome in terms of GC-content, codon usage and gene functions. It also carries terminal inverted repeats identical to the chromosome. EEC1 is enriched in biosynthetic gene clusters (BGCs), including the only copy of the BGCs for the spore pigment and the surfactant peptide SapB, metabolites essential for the organism's lifecycle. EEC1 contains an origin of replication with at least some chromosomal properties, and its replication is likely to depend on functions provided by chromosomally located genes. Further comparison of Embleya spp. genomes suggests that EEC1-like replicons are conserved across the genus, in contrast to other known large linear extrachromosomal replicons (megaplasmids) in the order. EEC1 is thus a hallmark of the Embleya genus and is central to its evolution within the Streptomycetaceae family. We propose EEC1 as a secondary chromosome, distinct from previously described secondary chromosomes that utilise plasmid-like replication mechanisms (chromids) and the largest secondary replicon reported in bacteria, to date. ### Competing Interest Statement Ernest Lacey is a Founder, Board Member, and the Managing Director of Microbial Screening Technology Pty. Ltd. The authors declare no competing financial interests. Biotechnology and Biological Sciences Research Council, https://ror.org/00cwqg982, BB/P021506/1, BBS/E/J/000PR9790, BB/X01097X/1, BB/M011216/1 Novo Nordisk Foundation, https://ror.org/04txyc737, NNF22OC0078997
www.biorxiv.org
Thom Booth
@thombooth.bsky.social
· Jun 29