Marco Trujillo
@trujillolab.bsky.social
440 followers
560 following
23 posts
Fascinated by ubiquitination. Works at the RWTH Aachen University. Tweets are my own.
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Marco Trujillo
Karla Feijs-Žaja
@kfeijszaja.bsky.social
· Apr 25
Mike Cohen
@michaelnadbio.bsky.social
· Apr 24

Modifying the modifiers: ubiquitination of ADP-ribosylation in human cells
Ubiquitination and ADP-ribosylation are protein post-translational modifications (PTMs)
which influence diverse protein properties. In vitro work has indicated that ubiquitin
can be ADP-ribosylated an...
www.cell.com
Reposted by Marco Trujillo
Reposted by Marco Trujillo
Reposted by Marco Trujillo
Jane Dudley
@janedudley.bsky.social
· Apr 11
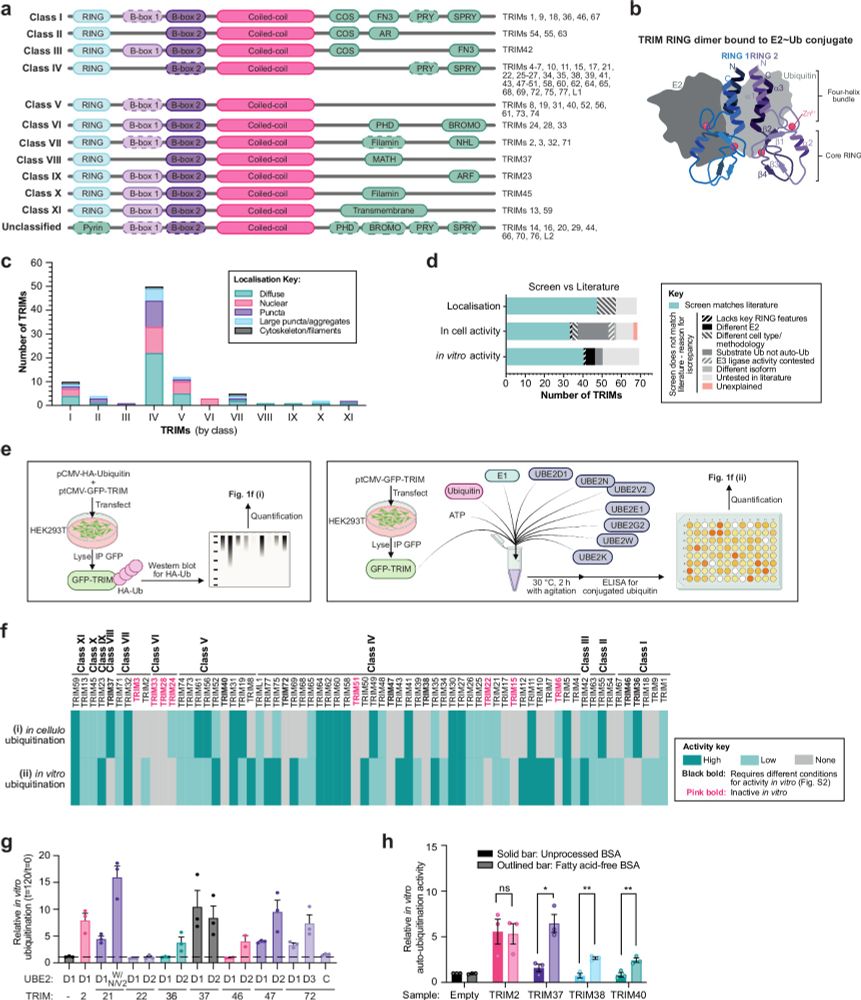
Identification of RING E3 pseudoligases in the TRIM protein family
Nature Communications - Understanding the catalytic and regulatory mechanisms of the ubiquitin system is key to tapping into its therapeutic potential in diseases where its dysfunction is...
rdcu.be
Reposted by Marco Trujillo
Marco Trujillo
@trujillolab.bsky.social
· Apr 10
Target of Rapamycin (TOR): A Master Regulator in Plant Growth, Development, and Stress Responses | Annual Reviews
The target of rapamycin (TOR) is a central regulator of growth, development, and stress adaptation in plants. This review delves into the molecular intricacies of TOR signaling, highlighting its conse...
www.annualreviews.org
Reposted by Marco Trujillo
Reposted by Marco Trujillo
Anthony Leung
@leunglab.bsky.social
· Apr 8

Interferon-induced PARP14-mediated ADP-ribosylation in p62 bodies requires the ubiquitin-proteasome system | The EMBO Journal
imageimageInterferon induces the formation of ADP-ribosylation (ADPr)-enriched p62 bodies, requiring
PARP14 and p62. Unlike canonical p62 bodies, these ADPr-enriched structures are independent
of auto...
www.embopress.org
Reposted by Marco Trujillo
Reposted by Marco Trujillo
Reposted by Marco Trujillo
Maddy Seale
@maddyseale.bsky.social
· Mar 30
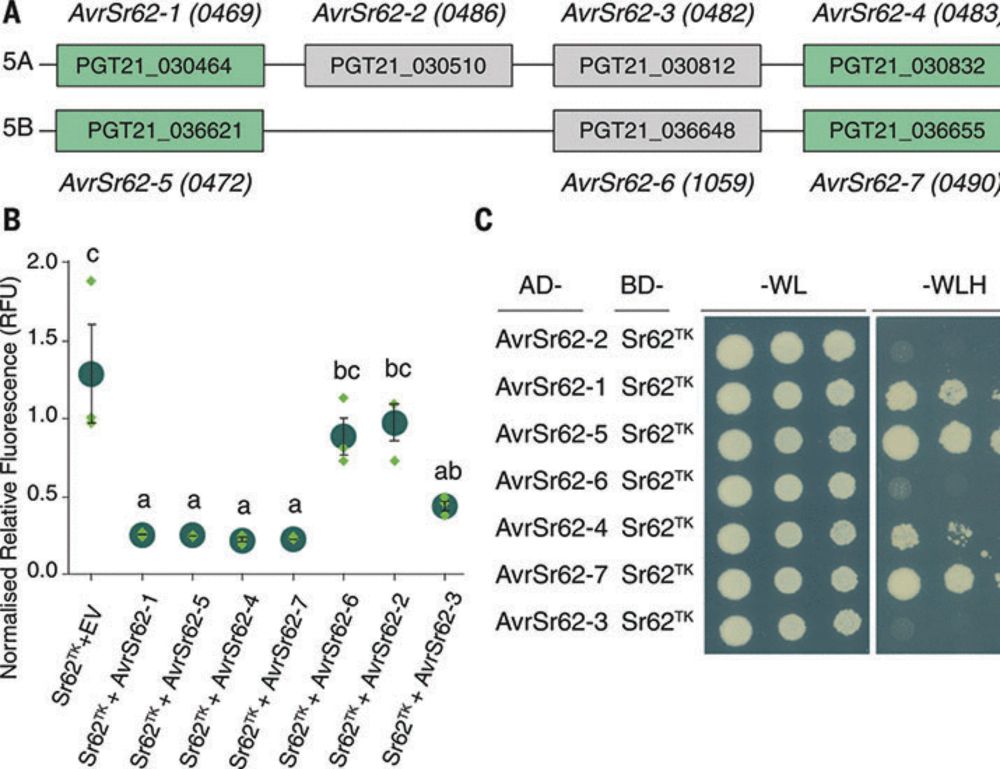
A wheat tandem kinase activates an NLR to trigger immunity
The role of nucleotide-binding leucine-rich repeat (NLR) receptors in plant immunity is well studied, but the function of a class of tandem kinases (TKs) that confer disease resistance in wheat and ba...
www.science.org
Reposted by Marco Trujillo
Micha Rapé Lab
@micharapelab.bsky.social
· Mar 27

Leucine aminopeptidase LyLAP enables lysosomal degradation of membrane proteins
Breakdown of every transmembrane protein trafficked to lysosomes requires proteolysis of their hydrophobic helical transmembrane domains. Combining lysosomal proteomics with functional genomic dataset...
www.science.org
Reposted by Marco Trujillo
Leo Kiss
@leokiss.bsky.social
· Mar 24
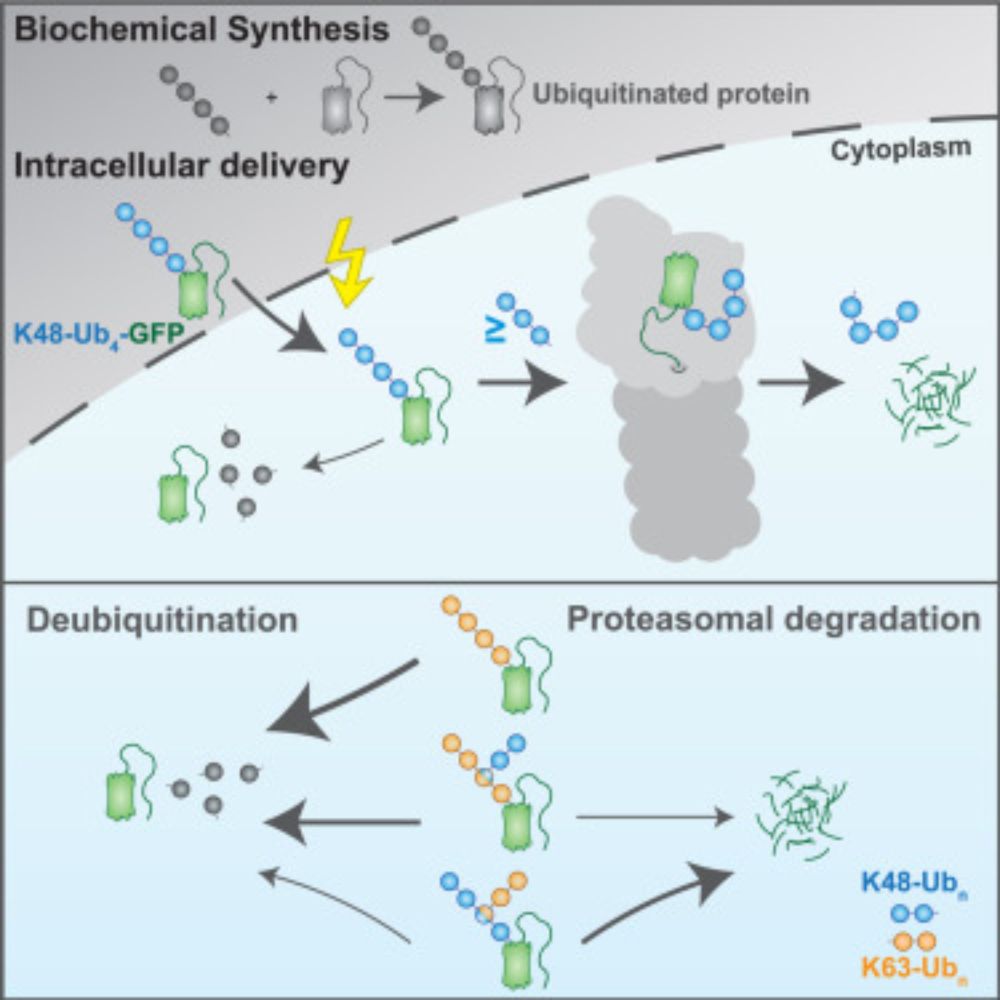
UbiREAD deciphers proteasomal degradation code of homotypic and branched K48 and K63 ubiquitin chains
Ubiquitin chains determine the fates of their modified proteins, including proteasomal
degradation. Kiss et al. present UbiREAD, a technology to monitor cellular degradation
and deubiquitination at hi...
www.cell.com