Read our paper @cp-cell.bsky.social!
❕Publication: doi.org/10.1016/j.ce...
❕Press Release: www.biochem.mpg.de/en/pressroom
@uoftmedicine.bsky.social
@erc.europa.eu #UPSmeetMet
Read our paper @cp-cell.bsky.social!
❕Publication: doi.org/10.1016/j.ce...
❕Press Release: www.biochem.mpg.de/en/pressroom
@uoftmedicine.bsky.social
@erc.europa.eu #UPSmeetMet
An abridged version of the method can also be found directly on the cryoET portal. Click "View All Info" on the page of one of the datasets contained in this deposition.

An abridged version of the method can also be found directly on the cryoET portal. Click "View All Info" on the page of one of the datasets contained in this deposition.
www.dementiaresearcher.nihr.ac.uk/job/research...

www.dementiaresearcher.nihr.ac.uk/job/research...
cryoetdataportal.czscience.com/depositions/...

cryoetdataportal.czscience.com/depositions/...
I’m proud to share this collaborative work using FIB milling and cryo-electron tomography to study tick-borne flavivirus replication. Thanks to all co-authors and collaborators — great teamwork #CryoET #ElectronTomography #Virology #Flavivirus #Microscopy #StructuralBiology
#virology #teamtomo
www.nature.com/articles/s41...

I’m proud to share this collaborative work using FIB milling and cryo-electron tomography to study tick-borne flavivirus replication. Thanks to all co-authors and collaborators — great teamwork #CryoET #ElectronTomography #Virology #Flavivirus #Microscopy #StructuralBiology
Register here: ter.li/zfiv48
#CellBiology #StructuralBiology #ElectronMicroscopy

Register here: ter.li/zfiv48
#CellBiology #StructuralBiology #ElectronMicroscopy
This hands-on workshop will introduce participants to standard image processing packages used to analyze cryo-ET data.
Apply by March 20: myumi.ch/lsi-cryo-wor...

This hands-on workshop will introduce participants to standard image processing packages used to analyze cryo-ET data.
Apply by March 20: myumi.ch/lsi-cryo-wor...
Using cryo-electron tomography, we show it forms amyloid structures inside lysosomes that mechanically rupture membranes – revealing a new paradigm for lysosomal failure.
🔗 doi.org/10.64898/202...
#CryoET

Using cryo-electron tomography, we show it forms amyloid structures inside lysosomes that mechanically rupture membranes – revealing a new paradigm for lysosomal failure.
🔗 doi.org/10.64898/202...
#CryoET
Source: https://phys.org/news/2026-01-tightening-focus-subcellular-snapshots-combined.html
Source: https://phys.org/news/2026-01-tightening-focus-subcellular-snapshots-combined.html

Segmentation of cryo-electron tomography data - https://ais-cryoet.readthedocs.org/ https://aiscryoet.org/
Author: mgflast
🏠Homepage
Segmentation of cryo-electron tomography data - https://ais-cryoet.readthedocs.org/ https://aiscryoet.org/
Author: mgflast
🏠Homepage
ter.li/of4cgh

ter.li/of4cgh

(1) PI3DETR: Parametric Instance Detection of 3D Point Cloud Edges With a Geometry-Aware 3DETR
(2) SemiETPicker: Fast and Label-Efficient Particle Picking for CryoET Tomography Using Semi-Supervised Learning
🔍 More at researchtrend.ai/communities/3DPC
#Biophysics #StructuralBiology #Organelles #CryoET

#Biophysics #StructuralBiology #Organelles #CryoET
Register here: buff.ly/wwNF6Zh
#SBGrid #StructuralBiology #Webinars

Register here: buff.ly/wwNF6Zh
#SBGrid #StructuralBiology #Webinars
#TeamTomo
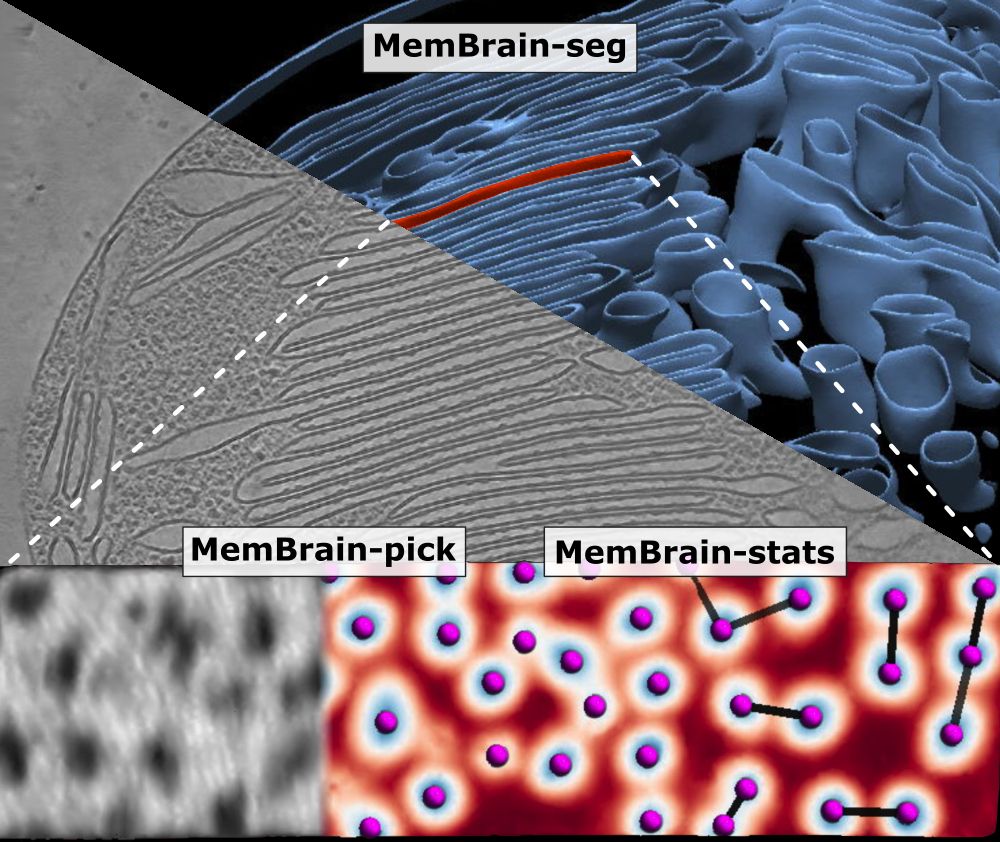
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
#TeamTomo

