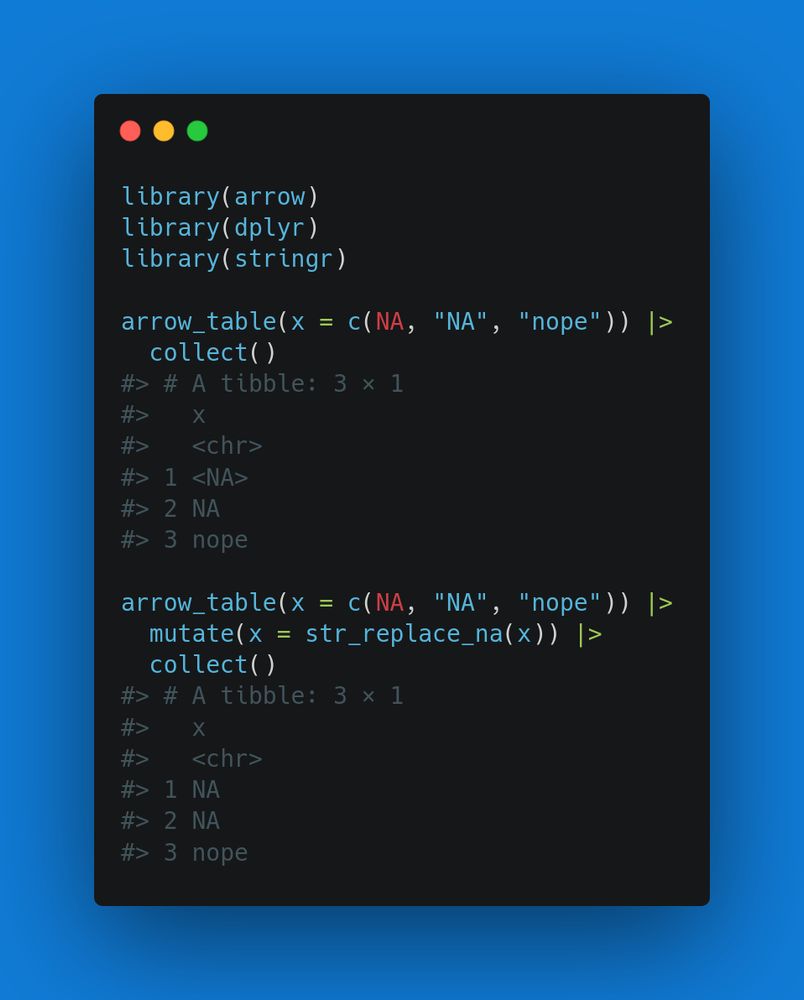

stringr::str_split(string, ",(?! )", simplify = TRUE)
stringr::str_split(string, ",(?! )", simplify = TRUE)

"Required Packages
library(quanteda)
library(quanteda.textstats)
library(quanteda.textplots)
library(readr)
library(dplyr)
library(ggplot2)
library(stringr)
library(DT)
library(tidytext)
Understanding Text Analytics Fundamentals
Text analytic...
Co..."
"Required Packages
library(quanteda)
library(quanteda.textstats)
library(quanteda.textplots)
library(readr)
library(dplyr)
library(ggplot2)
library(stringr)
library(DT)
library(tidytext)
Understanding Text Analytics Fundamentals
Text analytic...
Co..."
stringr.tidyverse.org/articles/fro...

stringr.tidyverse.org/articles/fro...
stringr::str_sort(x, numeric = TRUE)
[1] "1" "8" "9A" "10" "21A" "21B" "40"
stringr::str_sort(x, numeric = TRUE)
[1] "1" "8" "9A" "10" "21A" "21B" "40"
x <- c("8", "10", "1", "40")
str_sort(x, numeric = TRUE)
[1] "1" "8" "10" "40"
x <- c("8", "10", "1", "40")
str_sort(x, numeric = TRUE)
[1] "1" "8" "10" "40"
Key takeaways:
awk = lightweight, precise
csvtk = robust, CSV-aware
stringr = complex logic in R
Always validate after replacing. Mistakes here can cost you a week.
Key takeaways:
awk = lightweight, precise
csvtk = robust, CSV-aware
stringr = complex logic in R
Always validate after replacing. Mistakes here can cost you a week.
Prefer R?
For more complex regex:
library(stringr)
df$V5 <- str_replace(df$V5, "pattern", "replacement")
Want to go further? Use str_replace_all() or mutate() from dplyr.
Prefer R?
For more complex regex:
library(stringr)
df$V5 <- str_replace(df$V5, "pattern", "replacement")
Want to go further? Use str_replace_all() or mutate() from dplyr.
con <- DBI::dbConnect(duckdb::duckdb())
dbplyr::translate_sql(stringr::str_trim(x), con = con)
#>
con <- DBI::dbConnect(duckdb::duckdb())
dbplyr::translate_sql(stringr::str_trim(x), con = con)
#>
Also thank you, I did not know about this function
Also thank you, I did not know about this function

Key takeaways:
• Regex cleans messy data fast
• Works on gene IDs, labels, metadata
• Combine with stringr for clarity
Key takeaways:
• Regex cleans messy data fast
• Works on gene IDs, labels, metadata
• Combine with stringr for clarity
cghlewis.github.io/data-wrangli...

![d12
# A tibble: 4 x 2
id item1
<dbl> <chr>
1 1 "c,a,p"
2 2 "a,p"
3 3 ""
4 4 "p"
Same as above, item1 represents student responses of a word. Each letter represents a correct phonetic sound. We want to count the number of correct sounds for each student. However, this time, our data has commas between letters so we cannot use stringr::str_length() because it will count the commas, giving us incorrect scores.
Instead we use stringr::str_count() and add the argument pattern = and we add the regex pattern “[a-z]” which says to count any lowercase letter a to z.
d12 %>%
dplyr::mutate(item1_score = stringr::str_count(item1, pattern = "[a-z]"))
# A tibble: 4 x 3
id item1 item1_score
<dbl> <chr> <int>
1 1 "c,a,p" 3
2 2 "a,p" 2
3 3 "" 0
4 4 "p" 1](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:zxaobre6qygeskx6nr2ew6lu/bafkreihh7fq44agdetkgp4gqdmlrrtaeooxibijl6wwcvxhxb7rrvyxyvi@jpeg)
cghlewis.github.io/data-wrangli...
"The WiRe" about trying to catch {stringr} Bell and tidyverse-ing up BaltimoRe
"The WiRe" about trying to catch {stringr} Bell and tidyverse-ing up BaltimoRe


