

Let me know if you’d like to be added or know someone who should be.
go.bsky.app/E7zXGSF
#RNAmodifications
#Drosophila
#Drosophila
Free to read here: rdcu.be/eR4zx

Free to read here: rdcu.be/eR4zx




The fossil, more than 400 million years old, offers hints about the origin of one of the greatest partnerships in the history of life on Earth.
www.nhm.ac.uk/discover/new...

The fossil, more than 400 million years old, offers hints about the origin of one of the greatest partnerships in the history of life on Earth.
www.nhm.ac.uk/discover/new...
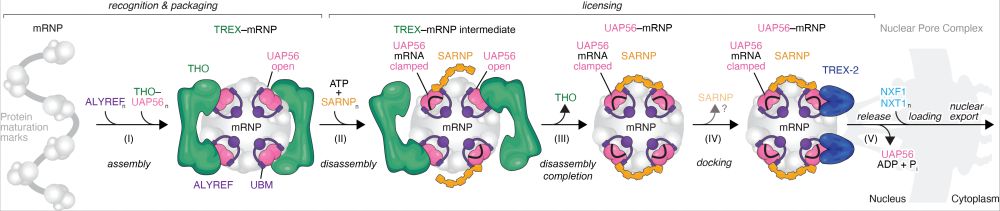
Very nice work from Holger Puchta & colleagues

Very nice work from Holger Puchta & colleagues



www.cell.com/cell/fulltex...
If you want to know more, read the 🧵 below:
www.cell.com/cell/fulltex...
If you want to know more, read the 🧵 below:
Our latest work from @immler.bsky.social lab 🐟🧬
Sex-specific responses of small RNAs and transposable elements to thermal stress in zebrafish germ cells
#TESky #TE #piRNA #miRNA #zebrafish #DanioDigest
🔗 Read here www.biorxiv.org/content/10.1...
A thread 🧵...

Our latest work from @immler.bsky.social lab 🐟🧬
Sex-specific responses of small RNAs and transposable elements to thermal stress in zebrafish germ cells
#TESky #TE #piRNA #miRNA #zebrafish #DanioDigest
🔗 Read here www.biorxiv.org/content/10.1...
A thread 🧵...


doi.org/10.1126/scie...

doi.org/10.1126/scie...

m6A-modified #EnhancerRNA & YTHDC1-BRD4 assemble transcriptional condensate
MINT-seq detects >3-fold more m6A peaks than MeRIP-seq
Stringtie annotate eRNAs & promoter upstream antisense RNAs, 24% & 48% are m6A+ (MCF7)
#MolCell 2021
www.cell.com/molecular-ce...

m6A-modified #EnhancerRNA & YTHDC1-BRD4 assemble transcriptional condensate
MINT-seq detects >3-fold more m6A peaks than MeRIP-seq
Stringtie annotate eRNAs & promoter upstream antisense RNAs, 24% & 48% are m6A+ (MCF7)
#MolCell 2021
www.cell.com/molecular-ce...
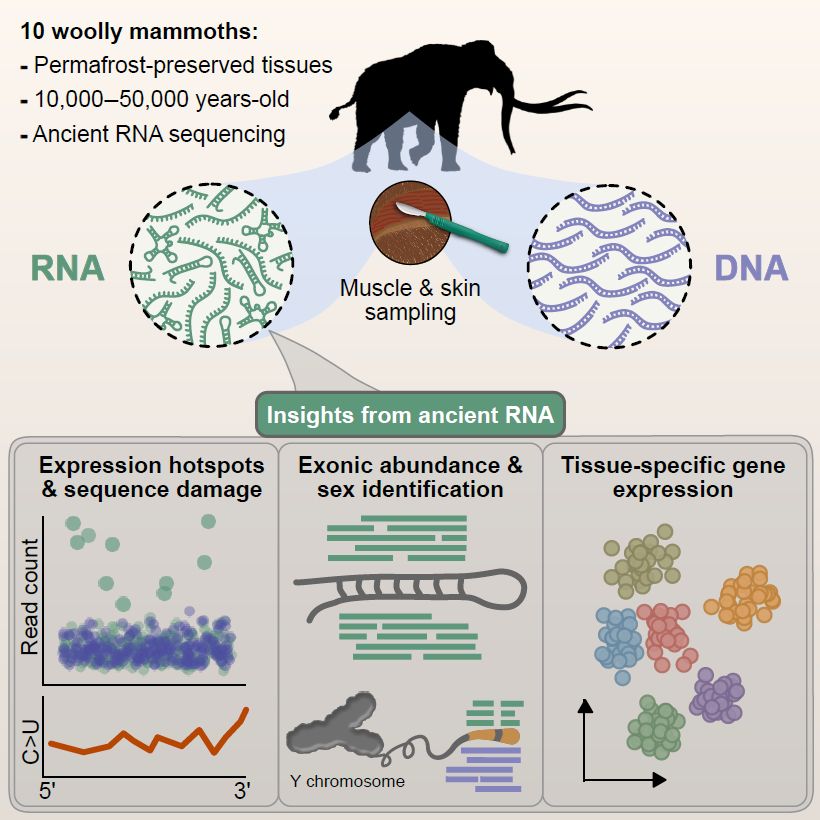
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


