
DeGrado, Arikin, Picotti (Keynotes). @lmkdassama.bsky.social @brianliau.bsky.social @rhodamine110.bsky.social @benlehner.bsky.social @alitavassoli.bsky.social
www.embl.org/about/info/c...
DeGrado, Arikin, Picotti (Keynotes). @lmkdassama.bsky.social @brianliau.bsky.social @rhodamine110.bsky.social @benlehner.bsky.social @alitavassoli.bsky.social
www.embl.org/about/info/c...
www.biorxiv.org/cgi/content/...

lnkd.in/dgsW4c3
Scientific Organizers: Britta Brügger, Robert Ernst, André Nadler, Christian Ungermann
Early registration & poster abstracts until January 31, 2026

lnkd.in/dgsW4c3
Scientific Organizers: Britta Brügger, Robert Ernst, André Nadler, Christian Ungermann
Early registration & poster abstracts until January 31, 2026
If you are interested in plasma membrane labeling, see the paper! 👇
pubs.rsc.org/en/content/a...
If you are interested in plasma membrane labeling, see the paper! 👇
pubs.rsc.org/en/content/a...
Also, we will have positions available next year. So stay tuned!
Find out which early-career researchers will receive funding this year, what they will be investigating, where they will be based... plus lots of other #ERCStG facts & figures for 2025!
➡️ buff.ly/IsafuFh
#FrontierResearch 🇪🇺#EUfunded #HorizonEurope
Also, we will have positions available next year. So stay tuned!
Read the #ScienceBooks Review: https://scim.ag/4mljVr1

Read the #ScienceBooks Review: https://scim.ag/4mljVr1
Huge thanks to Gaelen Guzman, a graduate student/postdoc in the lab who built it from scratch.
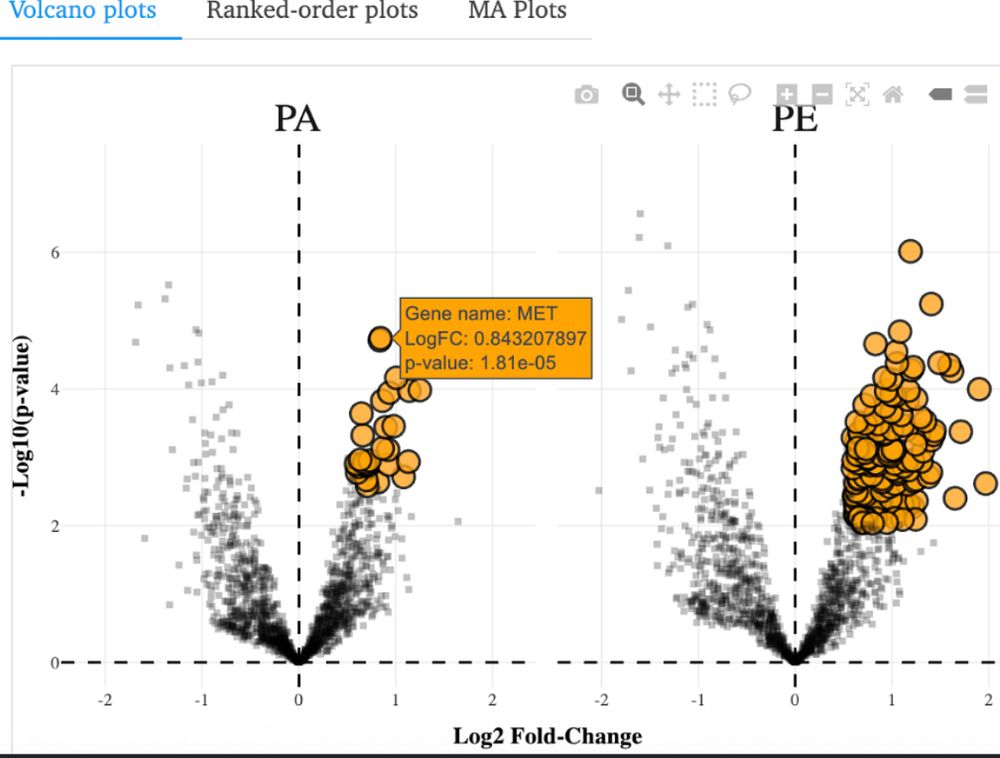
Huge thanks to Gaelen Guzman, a graduate student/postdoc in the lab who built it from scratch.
If you are looking for a postdoc position and you are interested in signaling lipids and proteomics, come join us in beautiful Switzerland!
More information on www.epfl.ch/labs/gr-schu...

If you are looking for a postdoc position and you are interested in signaling lipids and proteomics, come join us in beautiful Switzerland!
More information on www.epfl.ch/labs/gr-schu...

To help all scientists w/o graphic skills create clear, accessible, and truthful charts!
-> Out in @nature Cell Biology: rdcu.be/erwl4
#DataVisualization #PhD #SciComm
Thx for review @bethcimini.bsky.social + 2
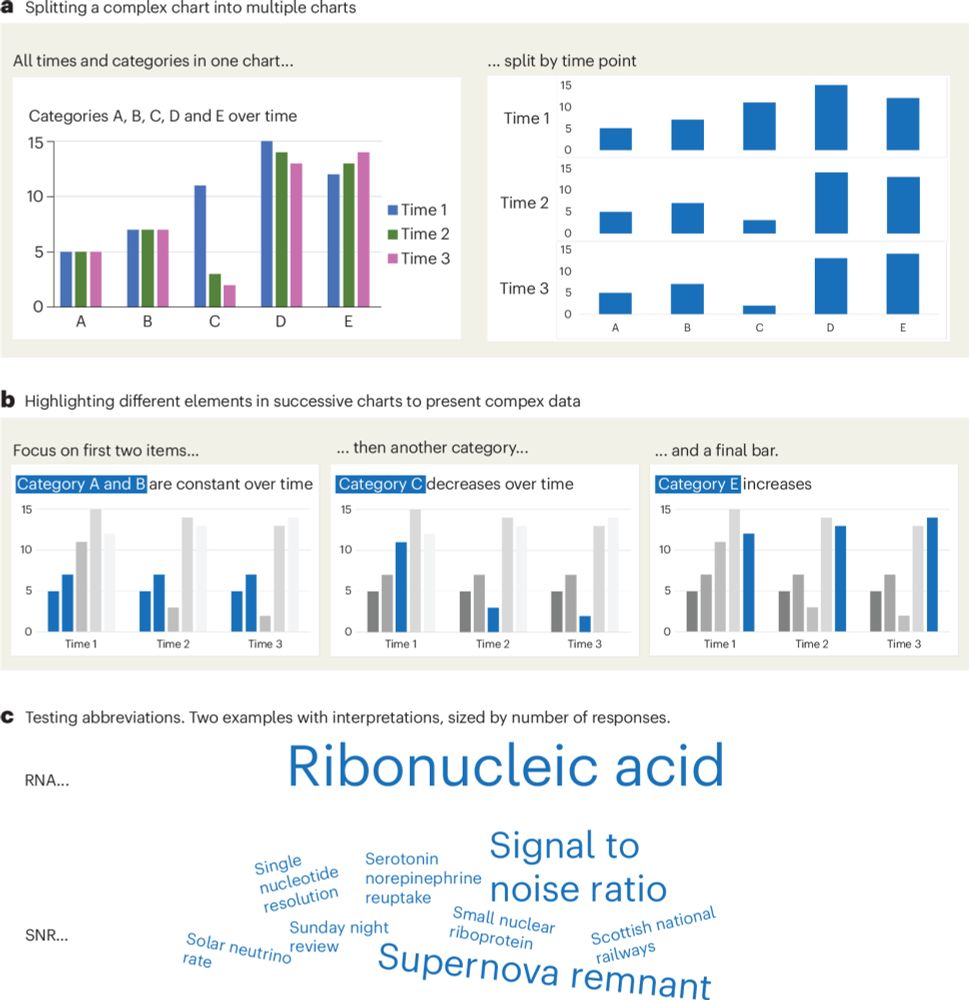
To help all scientists w/o graphic skills create clear, accessible, and truthful charts!
-> Out in @nature Cell Biology: rdcu.be/erwl4
#DataVisualization #PhD #SciComm
Thx for review @bethcimini.bsky.social + 2
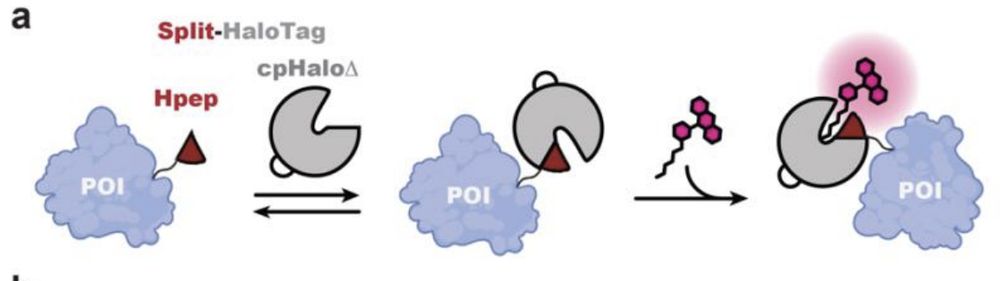
A new split Halotag system with higher affinity of the complements + lower background. The system works with our SiR-CA & CPY-CA halotag ligands and enables STED imaging or FLIM multiplexing.
The short 14 aa tag Hpep enables easy cloning free CRISPR-KI.

![Live-cell confocal imaging of histone H2B type-2E-Hpep11 CRISPR KI cell lines after 2-hours labeling with the specified CA- ligand [100 nM]. Images were taken with optimal image acquisition parameter for each dye. Scale bar: 50 μm.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:uf4m7zlmcjlg2biwn4s2k32y/bafkreibozn5fz6sgeksqrz3vmy6uiydtnvwmb5my7r3xecko2erokryn3a@jpeg)
![(a) Confocal laser scanning microscopy (CLSM) and STED images of mitochondria in U2OS cells coexpressing cpHaloΔ3 and TOM20-Hpep, either overexpressed or endogenously tagged. Scale bar: 1 μm. Pixel intensities scaled according to reference bar. (b) Representative CLSM, STED images of the CRISPR/Cas9 KI cells expressing TOM20 tagged with intact HaloTag (upper) or Hpep11 (bottom). (c) Intensity profiles along mitochondrial tubules (red and blue lines in b). Scale bar: 2 μm. (d) Representative CLSM and STED images of endogenous Hpep11-tagged clathrin with cpHaloΔ3 coexpression. Scale bar: 10 μm (overview) and 2 μm (magnification). (e) Representative CLSM, STED
images of endogenously tagged tubulin beta 4B with Hpep11. Scale bar: 10 μm (overview) and 2 μm (magnification). (f) Intensity profiles along tubulin filaments (red and blue lines in e) Means ± s.d. of the filament diameters were calculated as full width at half maximum (FWHM) from n=20 microtubule filaments, ≥ 2 images. A slight increase in cytosolic signal was noted in cells tagged with split-HaloTagat TUBB4B, compared to cells tagged with the full-length HaloTag, which may result from the presence of unbound but labeled cpHaloΔ3. All images were acquired after labeling with CA-SiR [100 nM] for one hour.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:uf4m7zlmcjlg2biwn4s2k32y/bafkreielty6fex37xy4caechbw5ebo3eypyiyvxxhkigeazd5gmy6tvlay@jpeg)
A new split Halotag system with higher affinity of the complements + lower background. The system works with our SiR-CA & CPY-CA halotag ligands and enables STED imaging or FLIM multiplexing.
The short 14 aa tag Hpep enables easy cloning free CRISPR-KI.
www.nature.com/articles/s41...
rdcu.be/epIB7
www.nature.com/articles/s41...
rdcu.be/epIB7
But maybe some of you are curious & 10% will feel seen. Or a little less alone. This isn’t about seeking sympathy.
It’s about sharing something hard to say out loud;
partly to heal, partly in case someone needs to hear it too. (1/3)
tinyurl.com/DudinO

But maybe some of you are curious & 10% will feel seen. Or a little less alone. This isn’t about seeking sympathy.
It’s about sharing something hard to say out loud;
partly to heal, partly in case someone needs to hear it too. (1/3)
tinyurl.com/DudinO

Check out the agenda with an amazing lineup of speakers and submit your abstract before 10 June 2025!
@embo.org
@biologists.bsky.social
meetings.embo.org/event/25-lip...

Check out the agenda with an amazing lineup of speakers and submit your abstract before 10 June 2025!
@embo.org
@biologists.bsky.social
meetings.embo.org/event/25-lip...





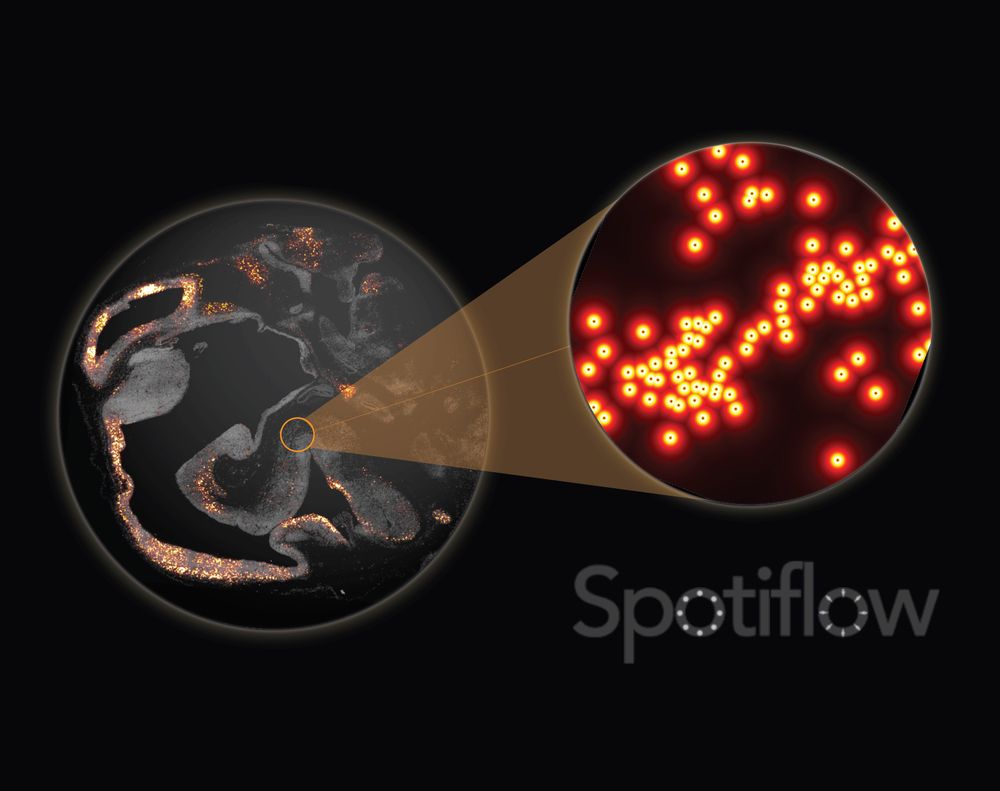
www.biorxiv.org/content/10.1...
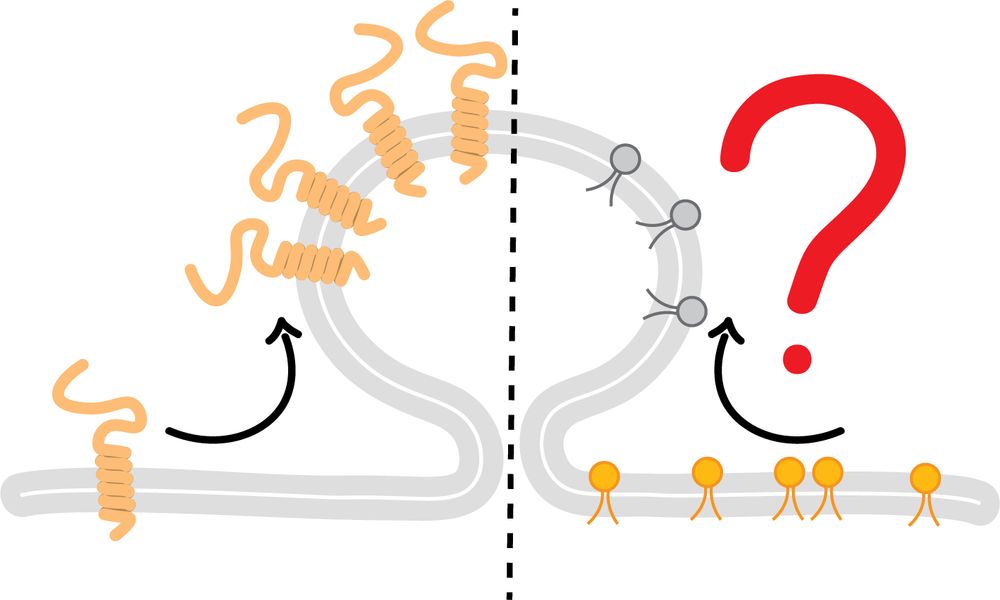
www.biorxiv.org/content/10.1...
SPY555-FastAct_X:
✅ Improved actin ligand captures very fast actin dynamics
✅ No toxicity
✅ High brightness
✅ Low phototoxicity
✅ High photoblueing & bleaching resistance
spirochrome.com/product/spy5...
Image: @lindawedemann.bsky.social from @mschuhmacher.bsky.social lab

SPY555-FastAct_X:
✅ Improved actin ligand captures very fast actin dynamics
✅ No toxicity
✅ High brightness
✅ Low phototoxicity
✅ High photoblueing & bleaching resistance
spirochrome.com/product/spy5...
Image: @lindawedemann.bsky.social from @mschuhmacher.bsky.social lab
Thank you @veselin-nasufovic.bsky.social

Thank you @veselin-nasufovic.bsky.social
www.biorxiv.org/content/10.1...
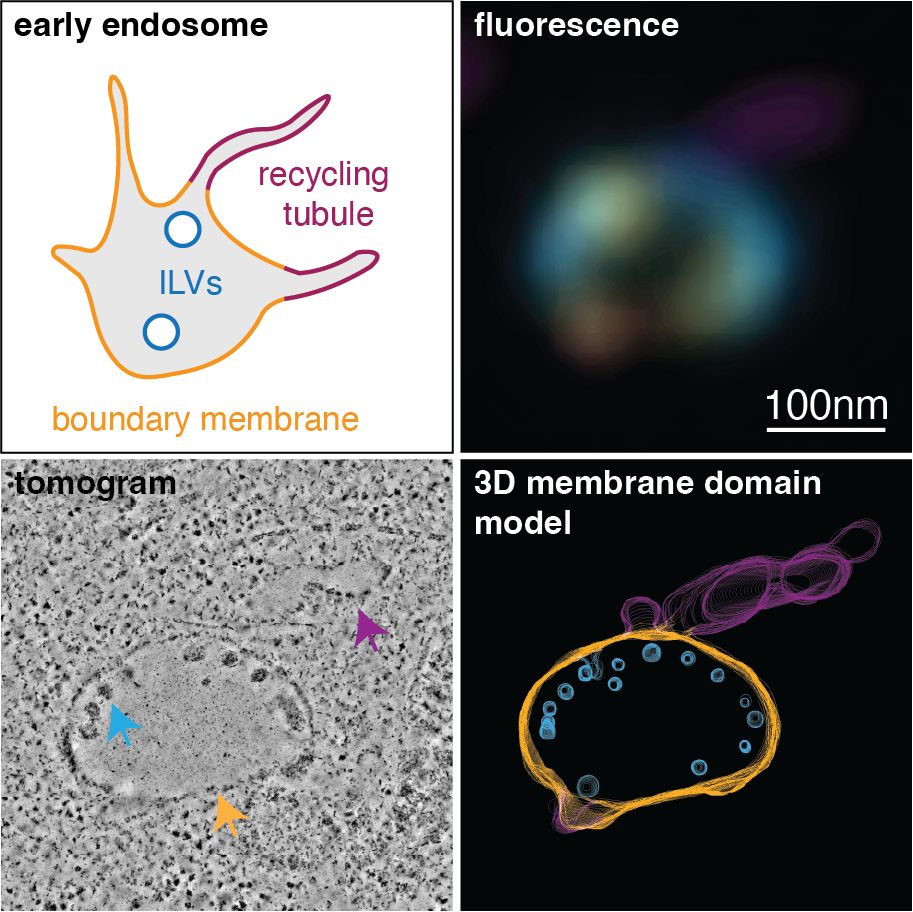
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Apply online 👇
stellenangebote.uni-marburg.de/jobposting/3...
📆Deadline 22.12 - please RT♥️🙏 #ChemBio

Apply online 👇
stellenangebote.uni-marburg.de/jobposting/3...
📆Deadline 22.12 - please RT♥️🙏 #ChemBio

