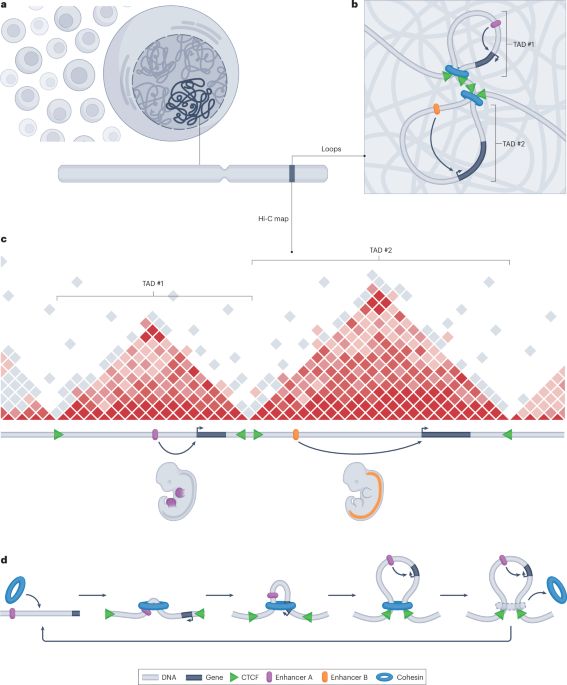
Yajie Zhu (PhD graduate) and Mariano Barbieri (postdoc, @spp2202.bsky.social Accelerator award) produced a package that will classify loops in your #3Dgenomics data based on (whichever) epigenomic data you have available. Keep reading...
1/n
www.biorxiv.org/content/10.6...
Yajie Zhu (PhD graduate) and Mariano Barbieri (postdoc, @spp2202.bsky.social Accelerator award) produced a package that will classify loops in your #3Dgenomics data based on (whichever) epigenomic data you have available. Keep reading...
1/n
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
go.nature.com/4pAbARf

go.nature.com/4pAbARf
#CellCycle #Biochemistry #Chromatin

#CellCycle #Biochemistry #Chromatin
www.nature.com/articles/s41...

www.nature.com/articles/s41...

www.nature.com/articles/s41...

www.nature.com/articles/s41...


academic.oup.com/nar/article/...

academic.oup.com/nar/article/...
#Epigenetics #Chromatin
---
Discover the breakthrough at epigenometech.com

#Epigenetics #Chromatin
---
Discover the breakthrough at epigenometech.com
Was co-submitted with @allanaschooley.bsky.social @jobdekker.bsky.social whose paper should also come out soon
Brief thread 👇
Was co-submitted with @allanaschooley.bsky.social @jobdekker.bsky.social whose paper should also come out soon
Brief thread 👇


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
👉https://rdcu.be/eHUxZ
bit.ly/424Iel9

👉https://rdcu.be/eHUxZ
bit.ly/424Iel9
By Jee Min Kim and Daniel Larson
➡️ https://tinyurl.com/gd352465
#transcription #molecularbiology #transcriptionbursting #embryonicdevelopment #stemcell #differentiation #mRNA #development #molecularmechanism
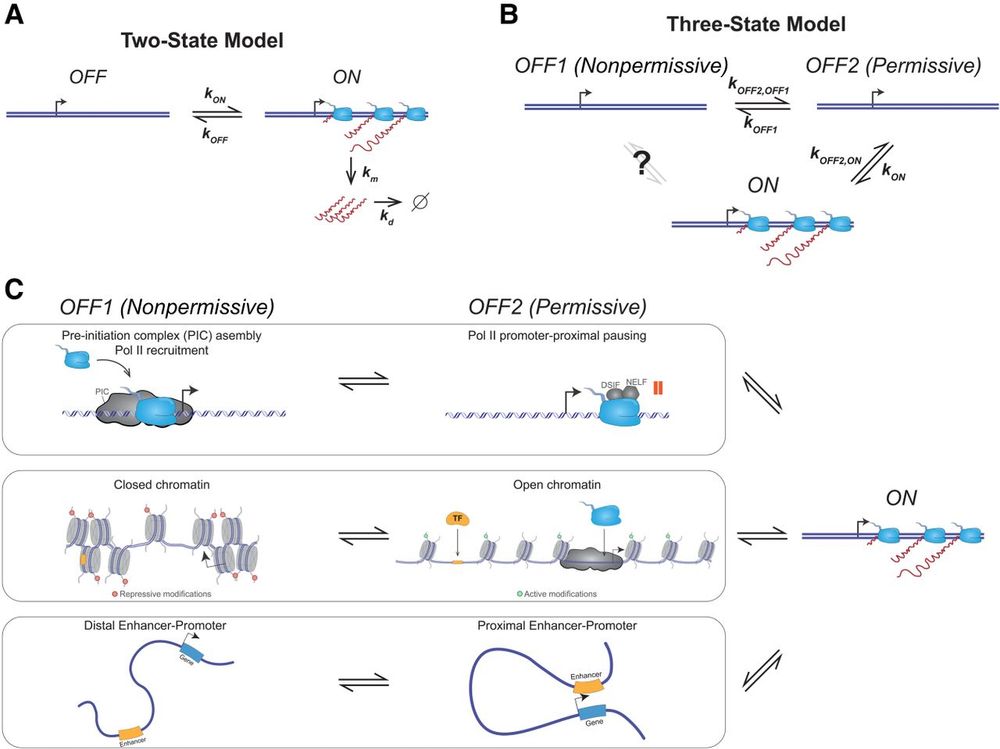
By Jee Min Kim and Daniel Larson
➡️ https://tinyurl.com/gd352465
#transcription #molecularbiology #transcriptionbursting #embryonicdevelopment #stemcell #differentiation #mRNA #development #molecularmechanism
Grateful to share my postdoctoral work introducing “tomographic & kinetically-enhanced DNA-PAINT” or in brief: tkPAINT. Out in @pnas.org!
doi.org/10.1073/pnas...
👇🧵

Grateful to share my postdoctoral work introducing “tomographic & kinetically-enhanced DNA-PAINT” or in brief: tkPAINT. Out in @pnas.org!
doi.org/10.1073/pnas...
👇🧵
STAG3-cohesin has a much shorter residence time which leads to altered 3D genome organization and STAG3-cohesin is important for male germ cell differentiation.
www.nature.com/articles/s41...

STAG3-cohesin has a much shorter residence time which leads to altered 3D genome organization and STAG3-cohesin is important for male germ cell differentiation.
www.nature.com/articles/s41...


www.biorxiv.org/content/10.1...
1/
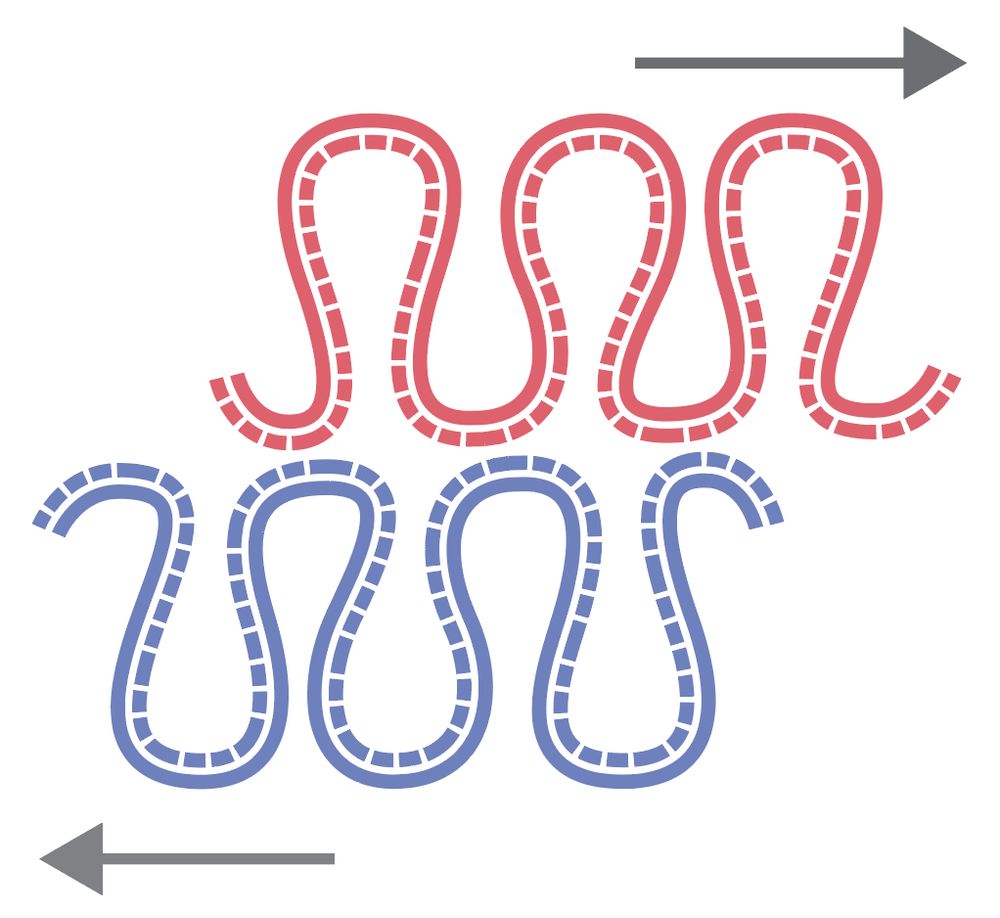
www.biorxiv.org/content/10.1...
1/