Chris McGann
@chrismcgann.bsky.social
160 followers
170 following
5 posts
Postdoctoral Research Fellow, Harvard Medical School & Dana-Farber Cancer Institute
Posts
Media
Videos
Starter Packs
Reposted by Chris McGann
Reposted by Chris McGann
Chris McGann
@chrismcgann.bsky.social
· Sep 6
Chris McGann
@chrismcgann.bsky.social
· Sep 6
Reposted by Chris McGann
Reposted by Chris McGann
Reposted by Chris McGann
Maitreya Dunham
@maitreya.bsky.social
· Jul 22

The yEvo Mutation Browser: Enhancing student understanding of experimental evolution and genomics through interactive data visualization
Experimental evolution is a powerful method for studying the relationship between genotype and phenotype by observing how populations genetically adapt to controlled selective pressures. In educationa...
www.biorxiv.org
Reposted by Chris McGann
Michael MacCoss
@maccoss.bsky.social
· Jul 2
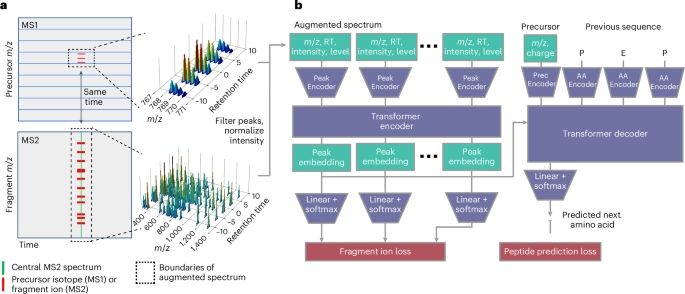
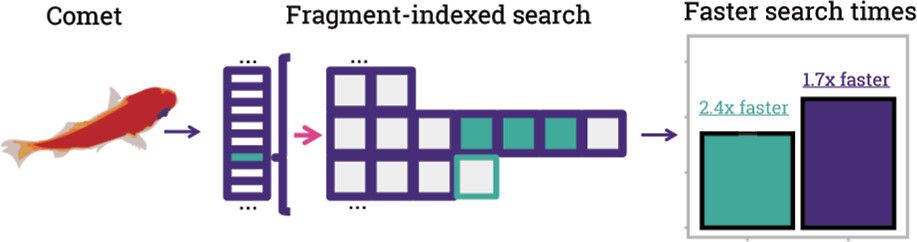
A transformer model for de novo sequencing of data-independent acquisition mass spectrometry data - Nature Methods
Cascadia is a mass spectrometry-based de novo sequencing model that uses a transformer architecture to handle data-independent acquisition data and achieves substantially improved performance across a...
www.nature.com
Reposted by Chris McGann
Reposted by Chris McGann
Reposted by Chris McGann
Nature Methods
@natmethods.nature.com
· Jun 16
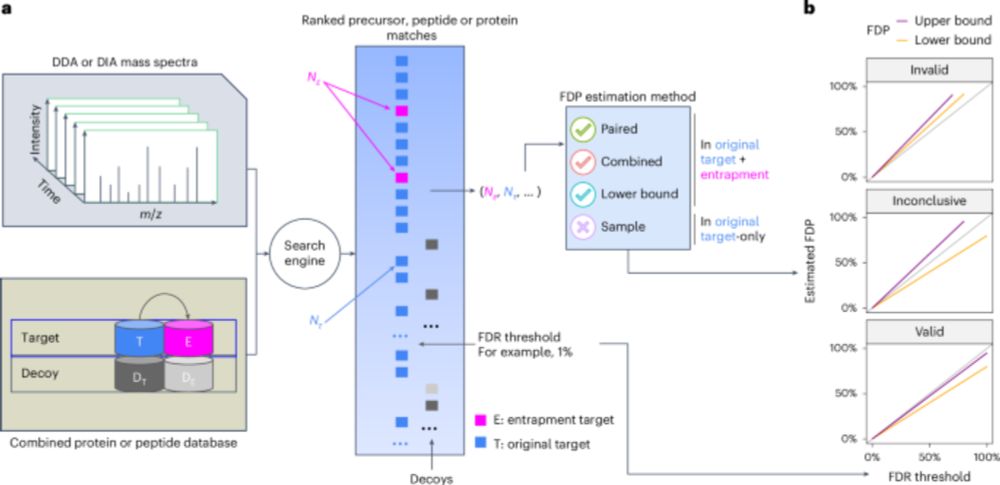
Assessment of false discovery rate control in tandem mass spectrometry analysis using entrapment - Nature Methods
A theoretical foundation for entrapment methods is presented, along with a method that enables more accurate evaluation of false discovery rate (FDR) control in proteomics mass spectrometry analysis p...
www.nature.com
Reposted by Chris McGann
Reposted by Chris McGann
Reposted by Chris McGann
Reposted by Chris McGann
Reposted by Chris McGann
UW Genome Sciences
@uwgenome.bsky.social
· Apr 22

A transformer model for de novo sequencing of data-independent acquisition mass spectrometry data
A core computational challenge in the analysis of mass spectrometry data is the de novo sequencing problem, in which the generating amino acid sequence is inferred directly from an observed fragmentat...
www.biorxiv.org
Reposted by Chris McGann
Reposted by Chris McGann
Reposted by Chris McGann