Dina Hochhauser
@dinahoch.bsky.social
430 followers
230 following
5 posts
PhD student @SorekLab @WeizmannScience studying phage-bacteria interactions 🦠, MGEs 🛒 & defence islands 🛡🏝
Posts
Media
Videos
Starter Packs
Pinned
Dina Hochhauser
@dinahoch.bsky.social
· Jul 29
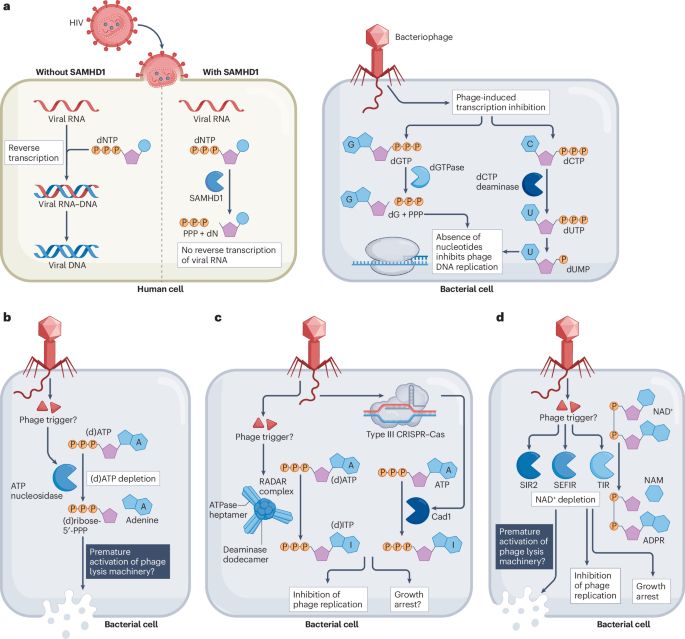
Manipulation of the nucleotide pool in human, bacterial and plant immunity - Nature Reviews Immunology
Modification of the nucleotide pool is emerging as key to innate immunity in animals, plants and bacteria. This Review explains how immune pathways conserved from bacteria to humans manipulate the nuc...
www.nature.com
Reposted by Dina Hochhauser
Reposted by Dina Hochhauser
Doudna Lab
@doudna-lab.bsky.social
· Aug 22
Erin Doherty
@erinedoherty.bsky.social
· Aug 22

Divergent viral phosphodiesterases for immune signaling evasion
Cyclic dinucleotides (CDNs) and other short oligonucleotides play fundamental roles in immune system activation in organisms ranging from bacteria to humans. In response, viruses use phosphodiesterase...
www.biorxiv.org
Reposted by Dina Hochhauser
Erin Doherty
@erinedoherty.bsky.social
· Aug 22

Divergent viral phosphodiesterases for immune signaling evasion
Cyclic dinucleotides (CDNs) and other short oligonucleotides play fundamental roles in immune system activation in organisms ranging from bacteria to humans. In response, viruses use phosphodiesterase...
www.biorxiv.org
Reposted by Dina Hochhauser
Kranzusch Lab
@kranzuschlab.bsky.social
· Aug 22

A widespread family of viral sponge proteins reveals specific inhibition of nucleotide signals in anti-phage defense
Chang et al. discover anti-CBASS 4 (Acb4), a family of viral sponges that inhibits
bacterial immunity by sequestering nucleotide immune signals. Acb4 homologs in phages
that infect hosts across all ma...
www.cell.com
Reposted by Dina Hochhauser
Vivek Mutalik
@vivekmutalik.bsky.social
· Aug 10
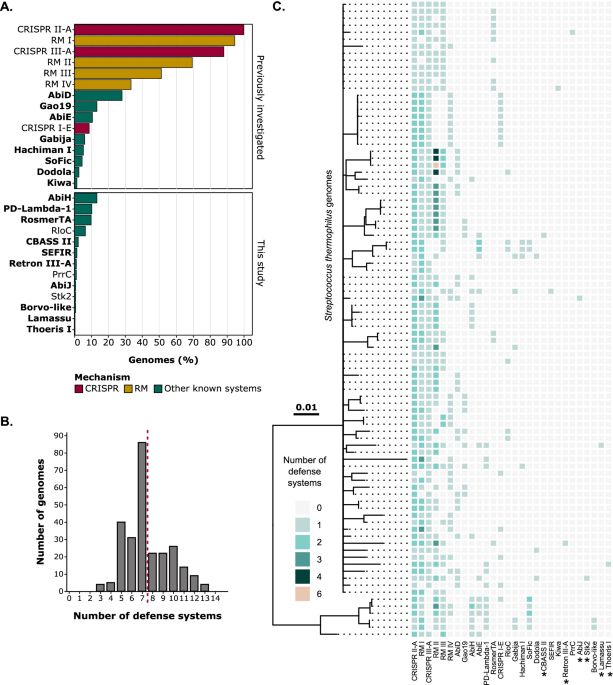
Strengthening phage resistance of Streptococcus thermophilus by leveraging complementary defense systems - Nature Communications
A study by Leprince et al. shows the effectiveness of Streptococcus thermophilus defense systems against dairy phages, their synergy with CRISPR-Cas, and the absence of fitness cost under lab or indus...
www.nature.com
Reposted by Dina Hochhauser
Tera Levin
@teralevin.bsky.social
· Aug 5

Hypermutable hotspot enables the rapid evolution of self/non-self recognition genes in Dictyostelium
Cells require highly polymorphic receptors to perform accurate self/non-self recognition. In the amoeba Dicytostelium discoideum, polymorphic TgrB1 & TgrC1 proteins are used to bind sister cells and e...
www.biorxiv.org
Dina Hochhauser
@dinahoch.bsky.social
· Aug 25
Romi Hadary
@romihadary.bsky.social
· Aug 25

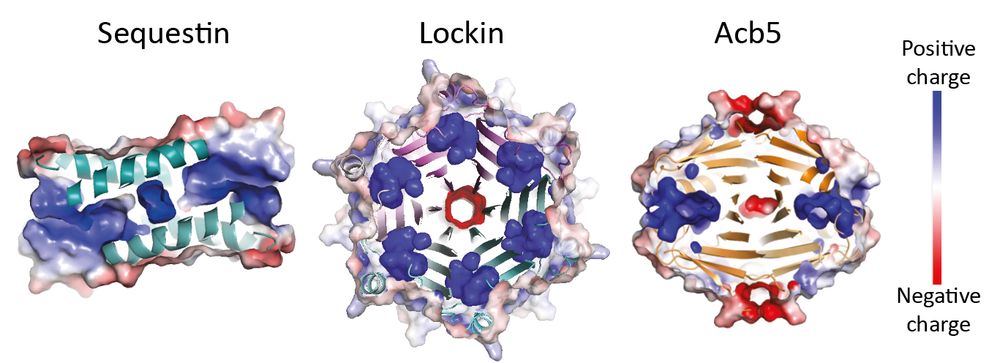
Functional diversity of phage sponge proteins that sequester host immune signals
Multiple bacterial immune systems, including CBASS, Thoeris, and Pycsar, employ signaling molecules that activate the immune response following phage infection. Phages counteract bacterial immune sign...
www.biorxiv.org
Reposted by Dina Hochhauser
Sorek Lab
@soreklab.bsky.social
· Jul 30
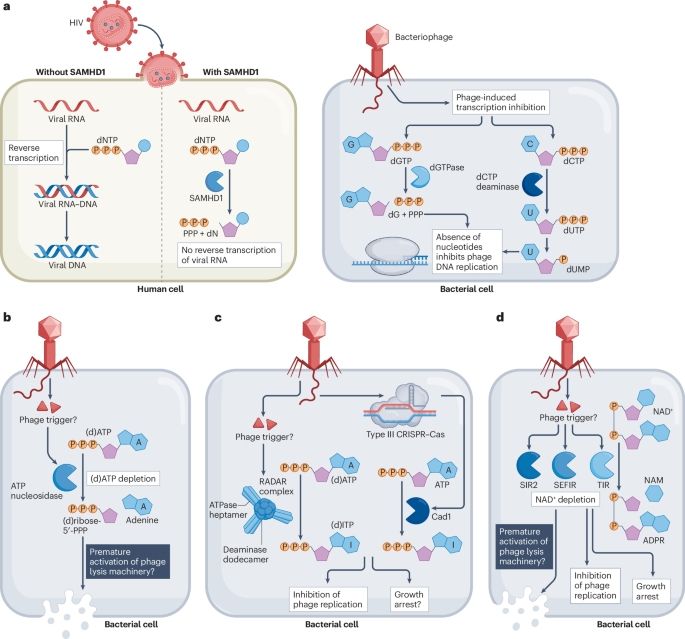
Manipulation of the nucleotide pool in human, bacterial and plant immunity - Nature Reviews Immunology
Modification of the nucleotide pool is emerging as key to innate immunity in animals, plants and bacteria. This Review explains how immune pathways conserved from bacteria to humans manipulate the nuc...
www.nature.com
Dina Hochhauser
@dinahoch.bsky.social
· Jul 29

Manipulation of the nucleotide pool in human, bacterial and plant immunity - Nature Reviews Immunology
Modification of the nucleotide pool is emerging as key to innate immunity in animals, plants and bacteria. This Review explains how immune pathways conserved from bacteria to humans manipulate the nuc...
www.nature.com
Reposted by Dina Hochhauser
Owen Tuck
@owentuck.bsky.social
· Jul 29

Recurrent acquisition of nuclease-protease pairs in antiviral immunity
Antiviral immune systems diversify by integrating new genes into existing pathways, creating new mechanisms of viral resistance. We identified genes encoding a predicted nuclease paired with a trypsin...
tinyurl.com
Reposted by Dina Hochhauser
Reposted by Dina Hochhauser
Erin Doherty
@erinedoherty.bsky.social
· Mar 31

A miniature CRISPR-Cas10 enzyme confers immunity by an inverse signaling pathway
Microbial and viral co-evolution has created immunity mechanisms involving oligonucleotide signaling that share mechanistic features with human anti-viral systems. In these pathways, including CBASS a...
www.biorxiv.org
Reposted by Dina Hochhauser
Reposted by Dina Hochhauser
Reposted by Dina Hochhauser
Reposted by Dina Hochhauser
Reposted by Dina Hochhauser
Sternberg Lab
@sternberglab.bsky.social
· Mar 26

Protein-primed DNA homopolymer synthesis by an antiviral reverse transcriptase
Bacteria defend themselves from viral predation using diverse immune systems, many of which sense and target foreign DNA for degradation. Defense-associated reverse transcriptase (DRT) systems provide...
www.biorxiv.org
Reposted by Dina Hochhauser
Reposted by Dina Hochhauser
Reposted by Dina Hochhauser
Reposted by Dina Hochhauser