Ed Chuong
@edchuong.bsky.social
1.8K followers
1.2K following
19 posts
Assistant Professor at the University of Colorado Boulder - Genome regulation, Transposons, Immunity - https://chuonglab.colorado.edu
Posts
Media
Videos
Starter Packs
Reposted by Ed Chuong
Reposted by Ed Chuong
Reposted by Ed Chuong
Reposted by Ed Chuong
Clément Canonne
@ccanonne.github.io
· Mar 20
Reposted by Ed Chuong
Reposted by Ed Chuong
Reposted by Ed Chuong
Ed Chuong
@edchuong.bsky.social
· Feb 5
Reposted by Ed Chuong
Ed Chuong
@edchuong.bsky.social
· Jan 10
Reposted by Ed Chuong
Ivan Zanoni
@lozanzi.bsky.social
· Dec 26
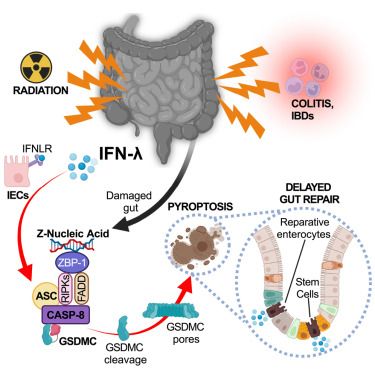
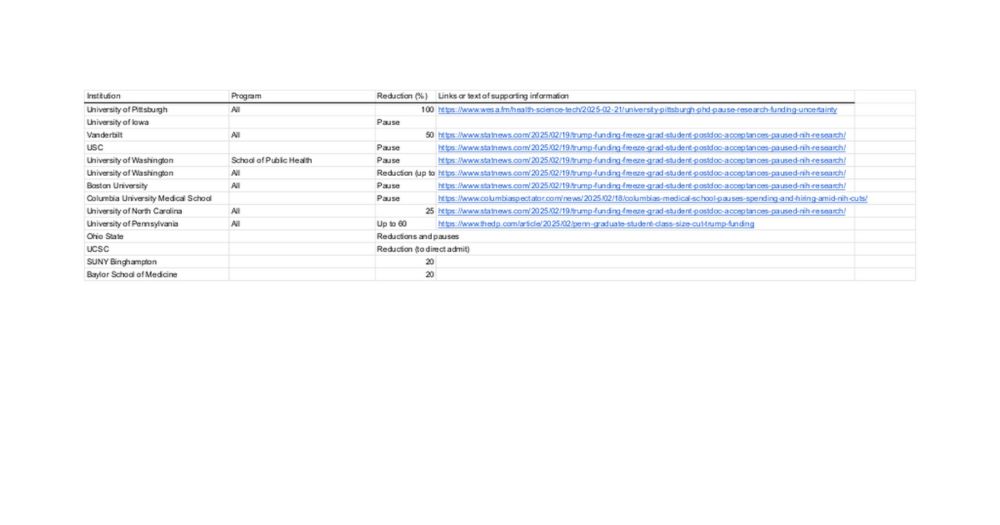
Type III interferons induce pyroptosis in gut epithelial cells and impair mucosal repair
Intestinal damage following colitis or irradiation induces type III interferons (IFNs), which delay the healing of the intestinal epithelium. IFN-λ directs the sensing of Z-form nucleic acids generated during gut injury and repair, triggering cell death and altering healthy epithelial regeneration.
www.cell.com
Reposted by Ed Chuong
Aaron Whiteley
@aaronwhiteley.bsky.social
· Dec 19
Emily Kibby
@emilykibby.bsky.social
· Dec 18

A bacterial NLR-related protein recognizes multiple unrelated phage triggers to sense infection
Immune systems must rapidly sense viral infections to initiate antiviral signaling and protect the host. Bacteria encode >100 distinct viral (phage) defense systems and each has evolved to sense cruci...
www.biorxiv.org
Reposted by Ed Chuong
Ed Chuong
@edchuong.bsky.social
· Dec 12

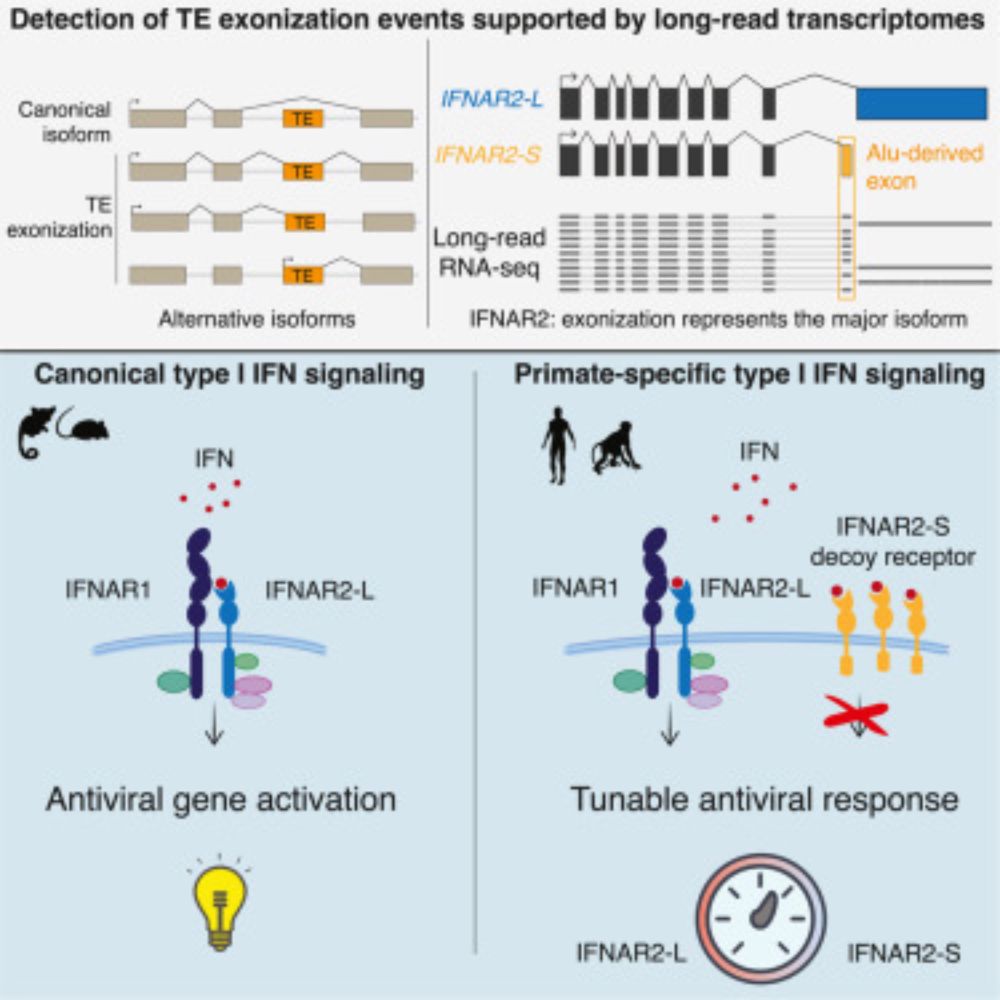
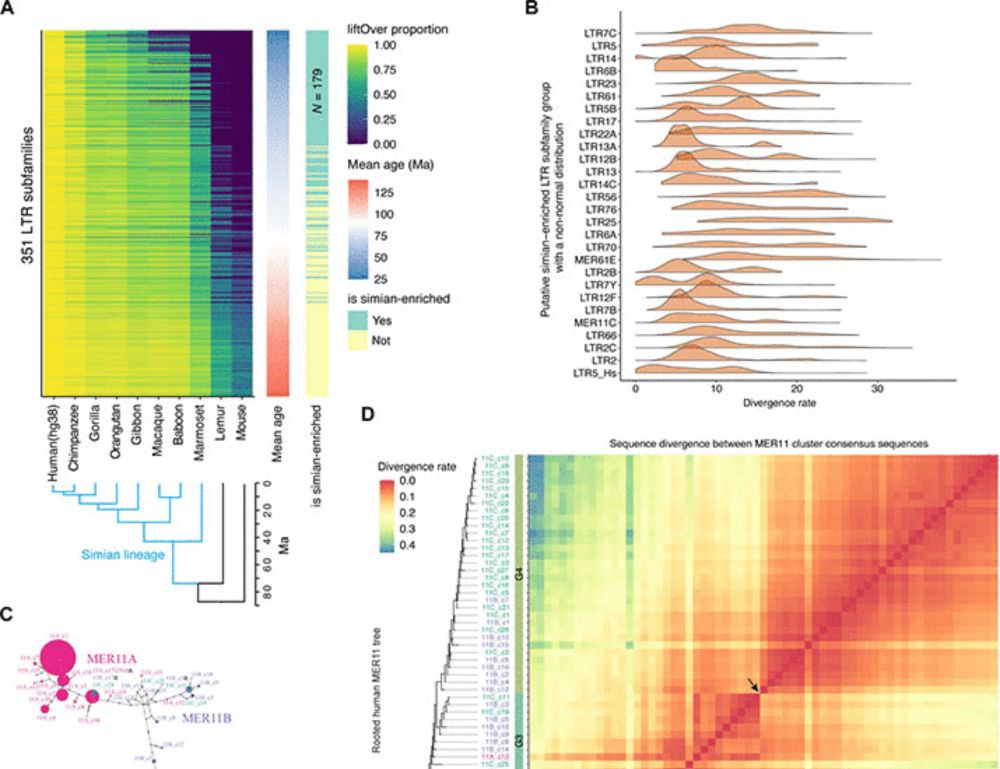
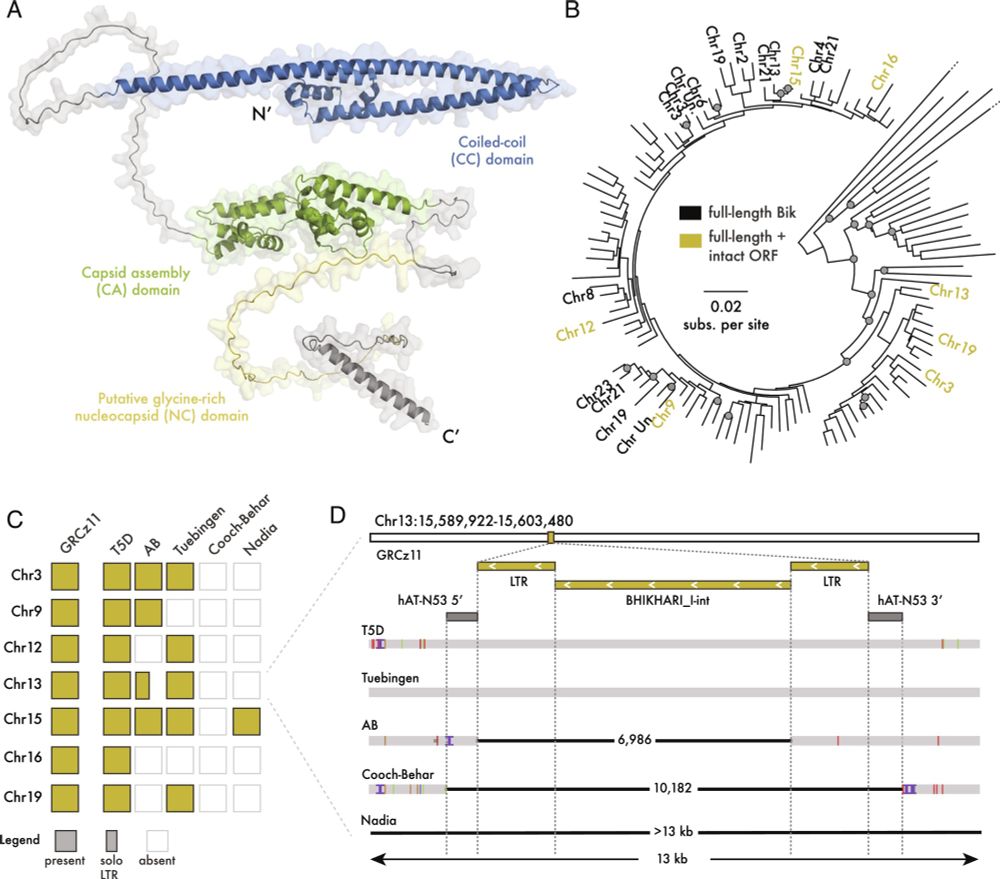
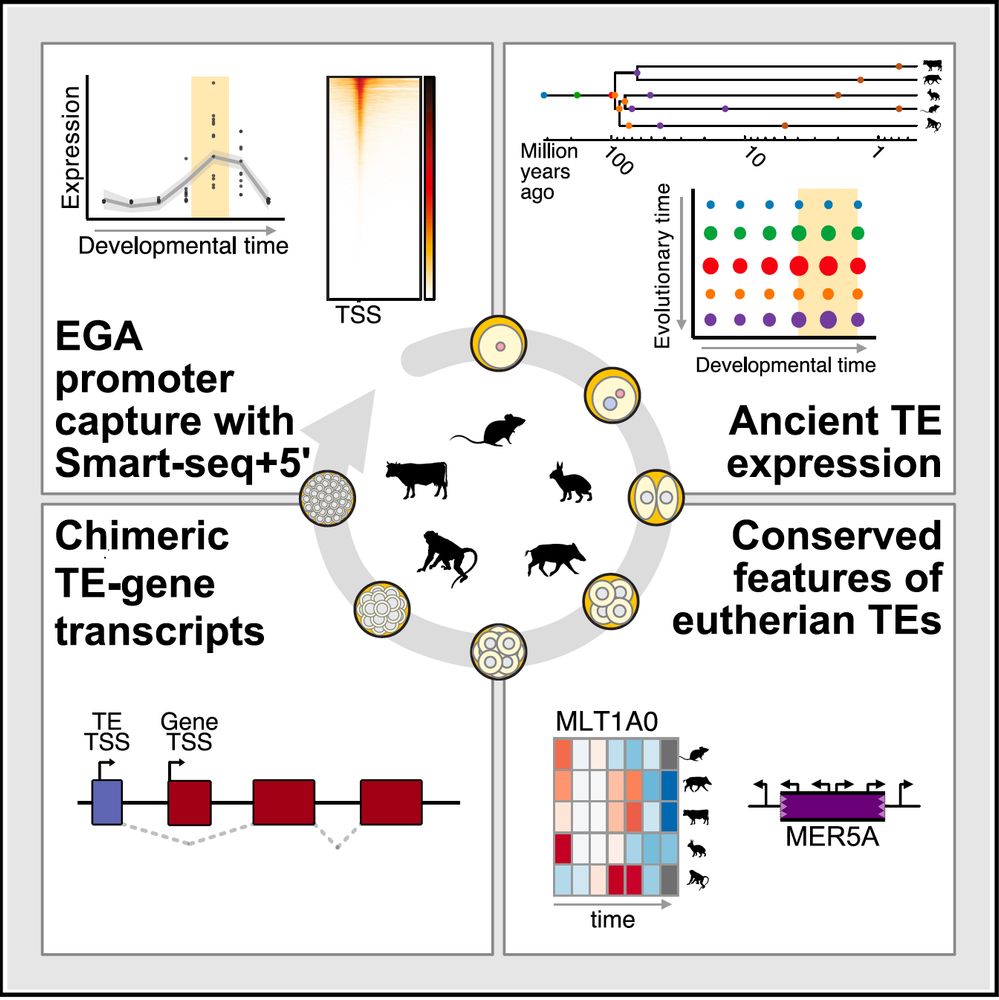
Transposable element exonization generates a reservoir of evolving and functional protein isoforms
Transposable element exonization by unannotated splicing events produces stable protein isoforms with acquired functions that are subject to evolutionary selection.
www.cell.com
Reposted by Ed Chuong
Ivan Zanoni
@lozanzi.bsky.social
· Dec 12

Transposable element exonization generates a reservoir of evolving and functional protein isoforms
Transposable element exonization by unannotated splicing events produces stable protein isoforms with acquired functions that are subject to evolutionary selection.
www.cell.com