Hsiu-Chuan Lin
@hsiuchuanlin.bsky.social
260 followers
260 following
25 posts
Group leader @crg.eu | Cell fate engineering and single-cell technology | A Taiwanese 🇹🇼
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Hsiu-Chuan Lin
Reposted by Hsiu-Chuan Lin
Reposted by Hsiu-Chuan Lin
Reposted by Hsiu-Chuan Lin
Reposted by Hsiu-Chuan Lin
Reposted by Hsiu-Chuan Lin
Stella Hurtley
@smhsci.bsky.social
· Aug 1
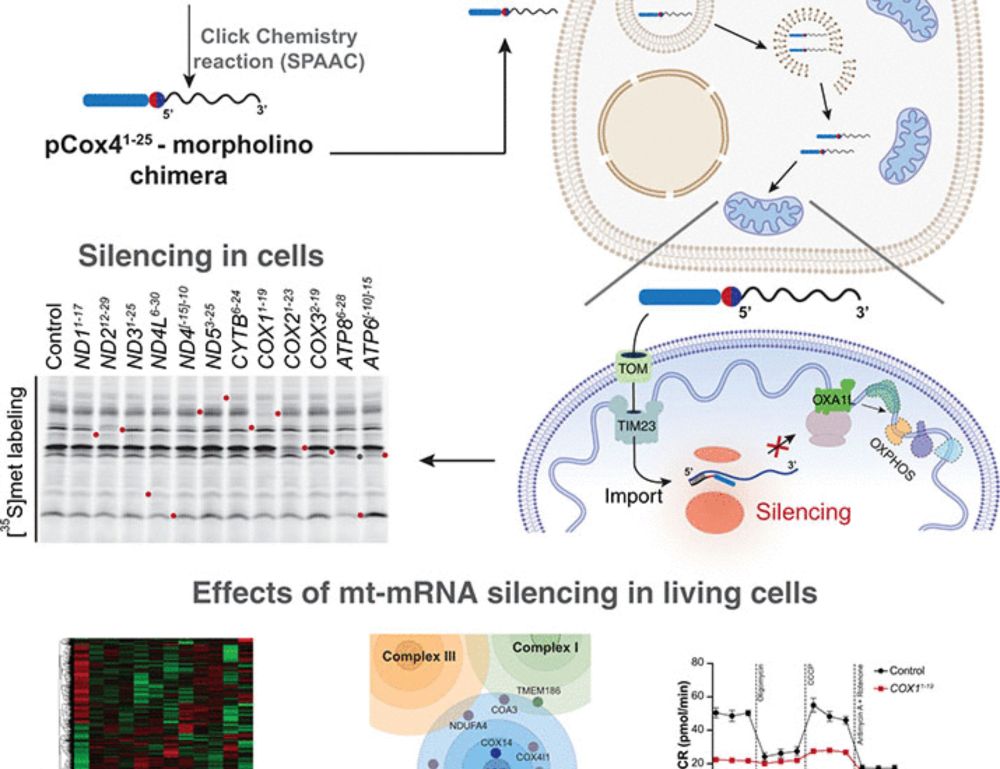
Silencing mitochondrial gene expression in living cells
Mitochondria fulfill central functions in metabolism and energy supply. They express their own genome, which encodes key subunits of the oxidative phosphorylation system. However, the central mechanis...
www.science.org
Reposted by Hsiu-Chuan Lin
Hsiu-Chuan Lin
@hsiuchuanlin.bsky.social
· Jul 15
Hsiu-Chuan Lin
@hsiuchuanlin.bsky.social
· Jul 15
Hsiu-Chuan Lin
@hsiuchuanlin.bsky.social
· Jul 15
Hsiu-Chuan Lin
@hsiuchuanlin.bsky.social
· Jul 15
Hsiu-Chuan Lin
@hsiuchuanlin.bsky.social
· Jul 12
Hsiu-Chuan Lin
@hsiuchuanlin.bsky.social
· Jul 12
Hsiu-Chuan Lin
@hsiuchuanlin.bsky.social
· Jul 12