Jon Markert
@jonmarkert.bsky.social
150 followers
86 following
11 posts
Postdoc Harvard Medical School, cryo-EM, chromatin, transcription.
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Lucas Farnung
@lucas.farnunglab.com
· Jan 30
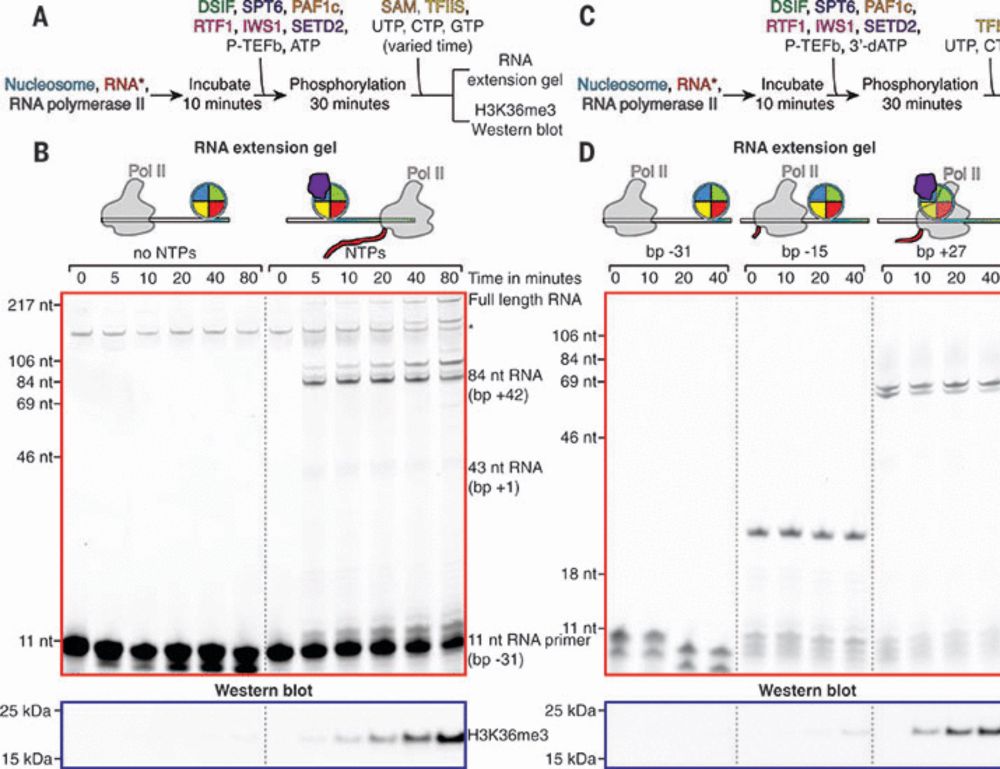
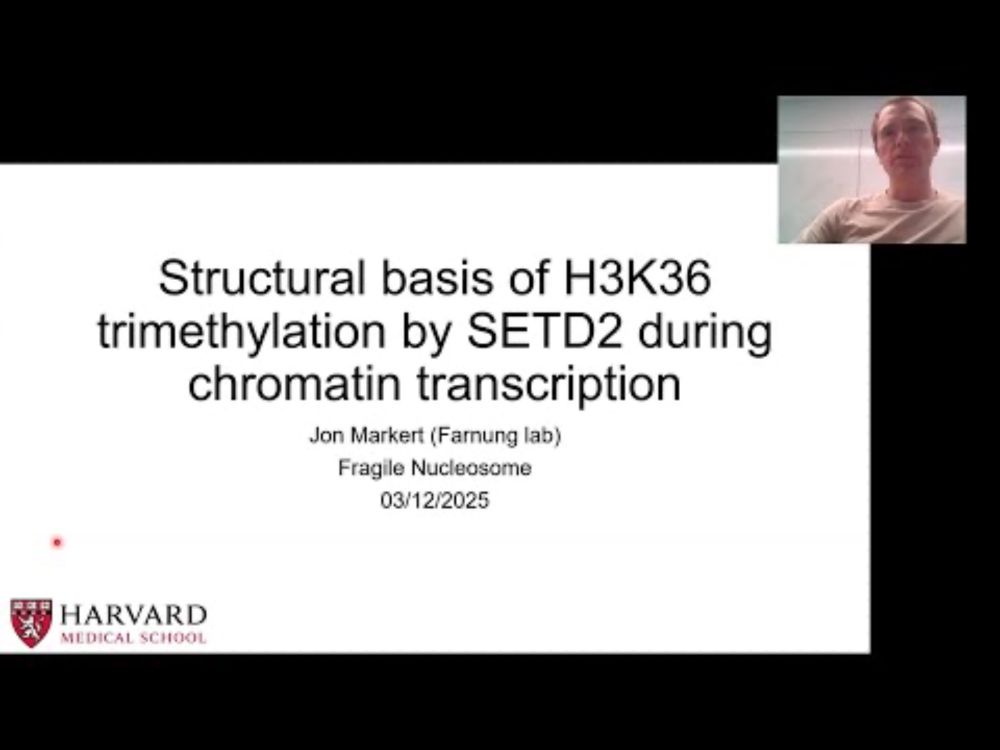
Structural basis of H3K36 trimethylation by SETD2 during chromatin transcription
During transcription, RNA polymerase II traverses through chromatin, and posttranslational modifications including histone methylations mark regions of active transcription. Histone protein H3 lysine ...
www.science.org
Reposted by Jon Markert
Seychelle Vos
@voslab.org
· Jan 1
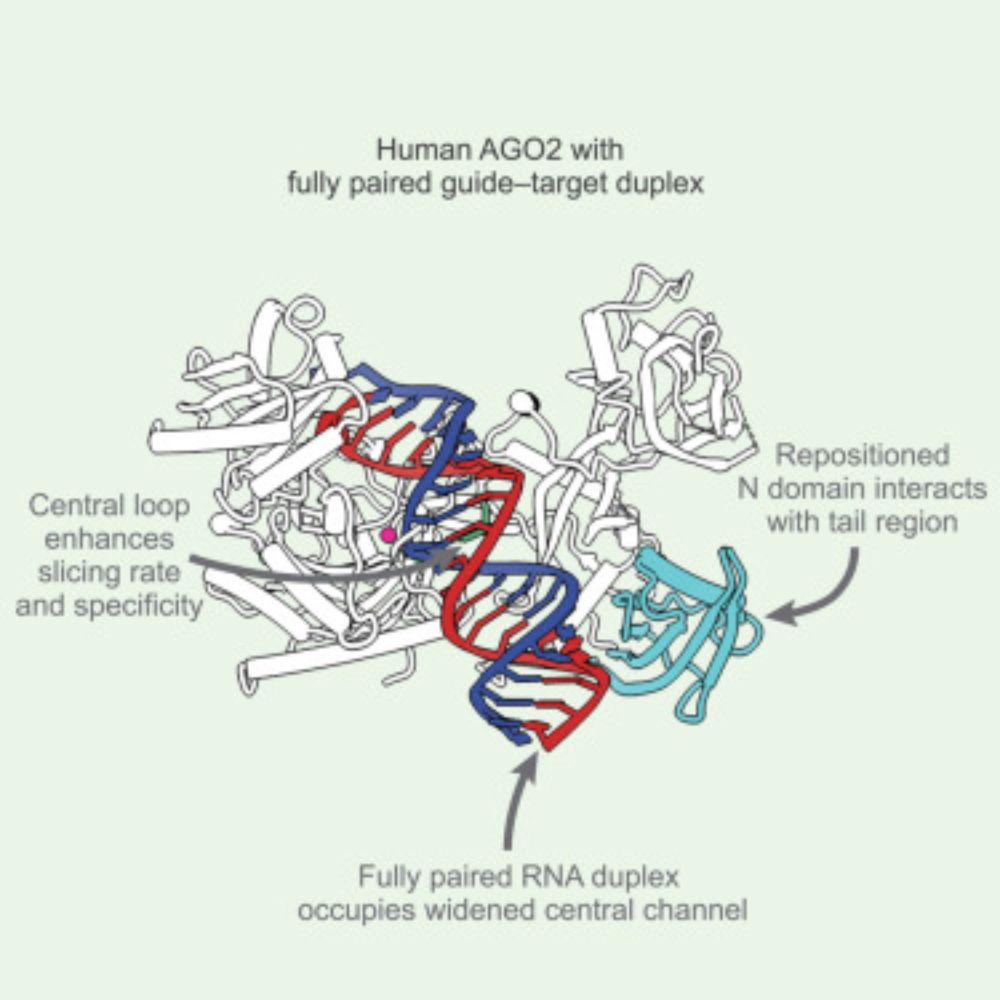
The structural basis for RNA slicing by human Argonaute2
Mohamed et al. report the cryoelectron microscopy structure of human AGO2 with fully paired guide RNA. Their analysis reveals the structural basis for the slicing activity that drives RNAi, showing that the slicing-competent conformation is achieved by domain movements and RNA-protein contacts distinct from those of conformational intermediates and prokaryotic homologs.
tinyurl.com
Reposted by Jon Markert
Reposted by Jon Markert
Reposted by Jon Markert
Di Jiang
@dijiang319.bsky.social
· Dec 13
Jon Markert
@jonmarkert.bsky.social
· Dec 13
Jon Markert
@jonmarkert.bsky.social
· Dec 13
Jon Markert
@jonmarkert.bsky.social
· Dec 12
Jon Markert
@jonmarkert.bsky.social
· Dec 12