Nikhil Milind
@nikhilmilind.dev
210 followers
410 following
15 posts
PhD Candidate in the Pritchard Lab at Stanford University. Interested in statistical and population genetics.
https://nikhilmilind.dev/
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Laura K Hayward
@lkhayward.bsky.social
· Aug 23
Reposted by Nikhil Milind
Matthew Aguirre
@aguirre404.bsky.social
· Aug 22
Reposted by Nikhil Milind
Roshni Patel
@roshnipatel.bsky.social
· Aug 6
Reposted by Nikhil Milind
Roshni Patel
@roshnipatel.bsky.social
· Aug 6
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Jonathan Pritchard
@jkpritch.bsky.social
· Jul 28
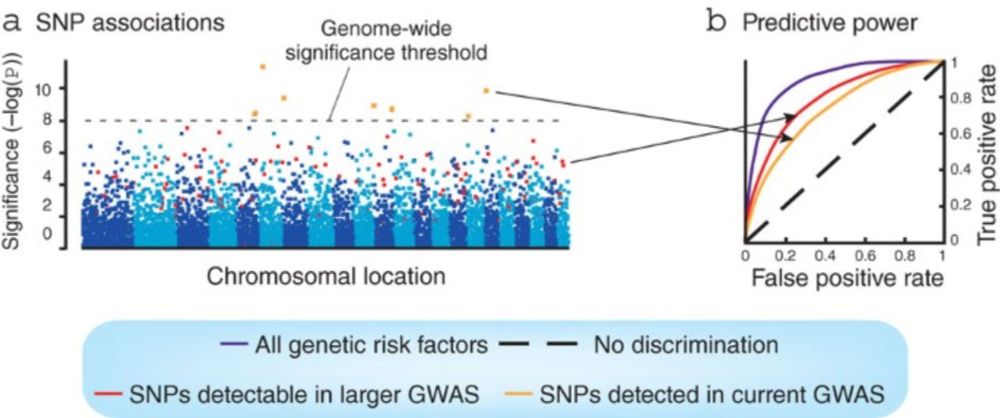
Hints of hidden heritability in GWAS - Nature Genetics
Although susceptibility loci identified through genome-wide association studies (GWAS) typically explain only a small proportion of the heritability, a classical quantitative genetic analysis now argu...
doi.org
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Hakhamanesh Mostafavi
@hakha.bsky.social
· Jul 25
Reposted by Nikhil Milind
Hanbin Lee
@epigenci.bsky.social
· Jul 19
Reposted by Nikhil Milind
Molly Przeworski
@mollyprz.bsky.social
· Jul 16
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Jeff Spence
@jeffspence.github.io
· Jun 2
Reposted by Nikhil Milind
Reposted by Nikhil Milind
Jeff Spence
@jeffspence.github.io
· May 30