Utz Ermel
@uermel.bsky.social
340 followers
680 following
14 posts
Scientist at Chan Zuckerberg Imaging Institute, Redwood City, CA
I like to make open source cryoET things.
Posts
Media
Videos
Starter Packs
Reposted by Utz Ermel
Reposted by Utz Ermel
Kyle Harrington
@kyleharrington.com
· Aug 26

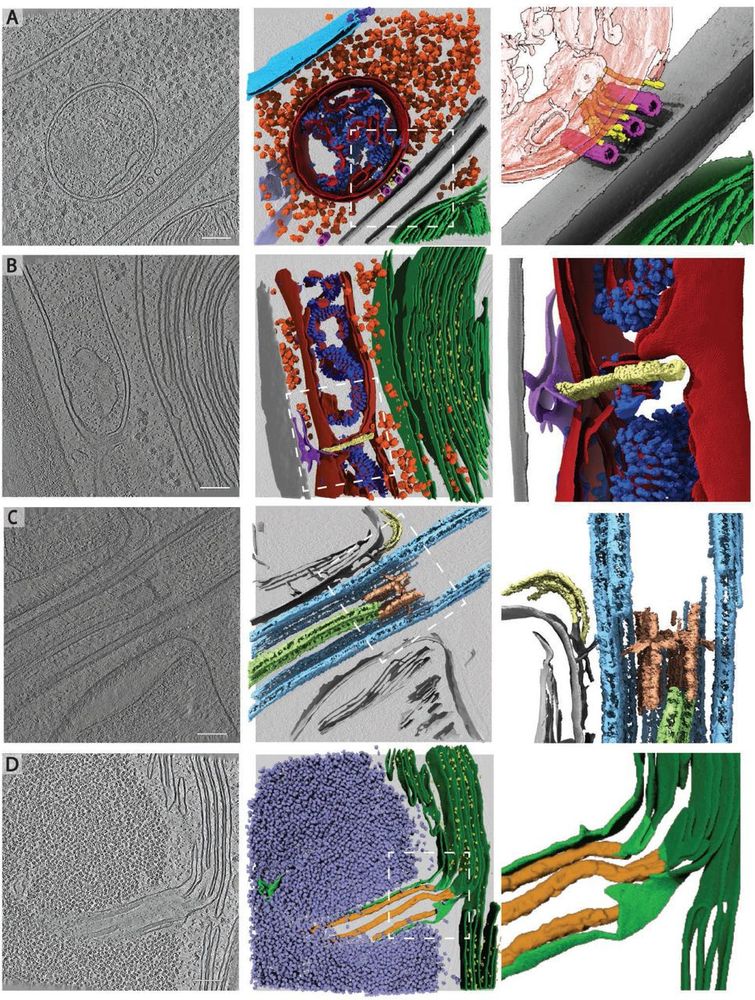
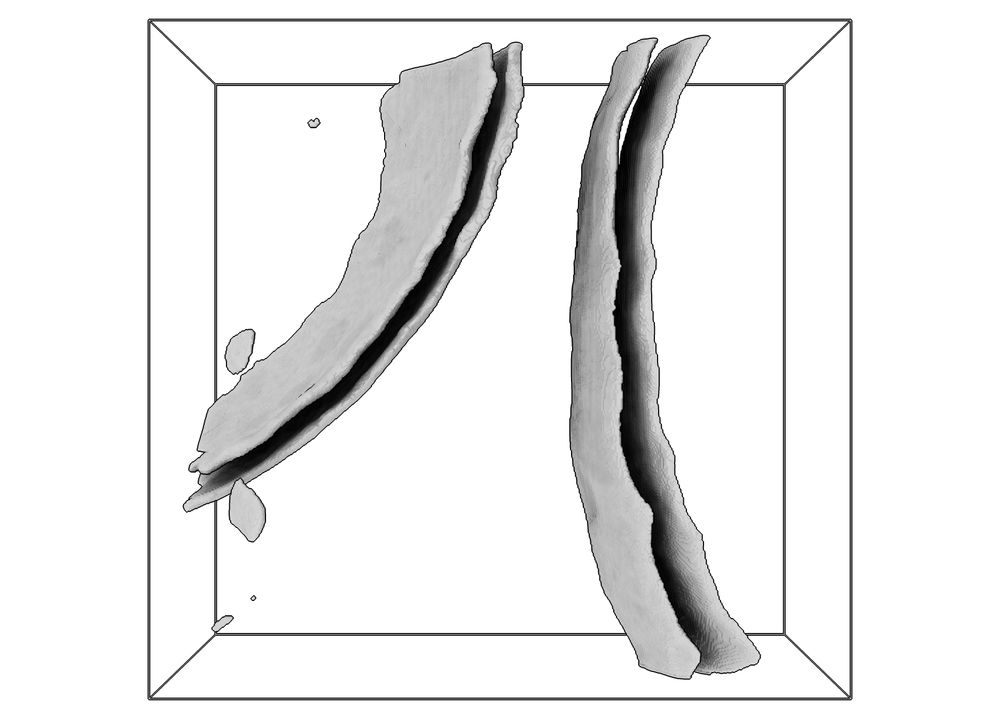
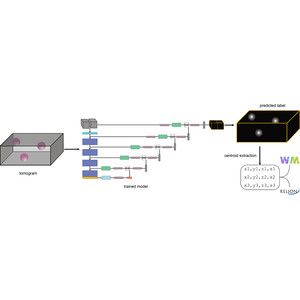
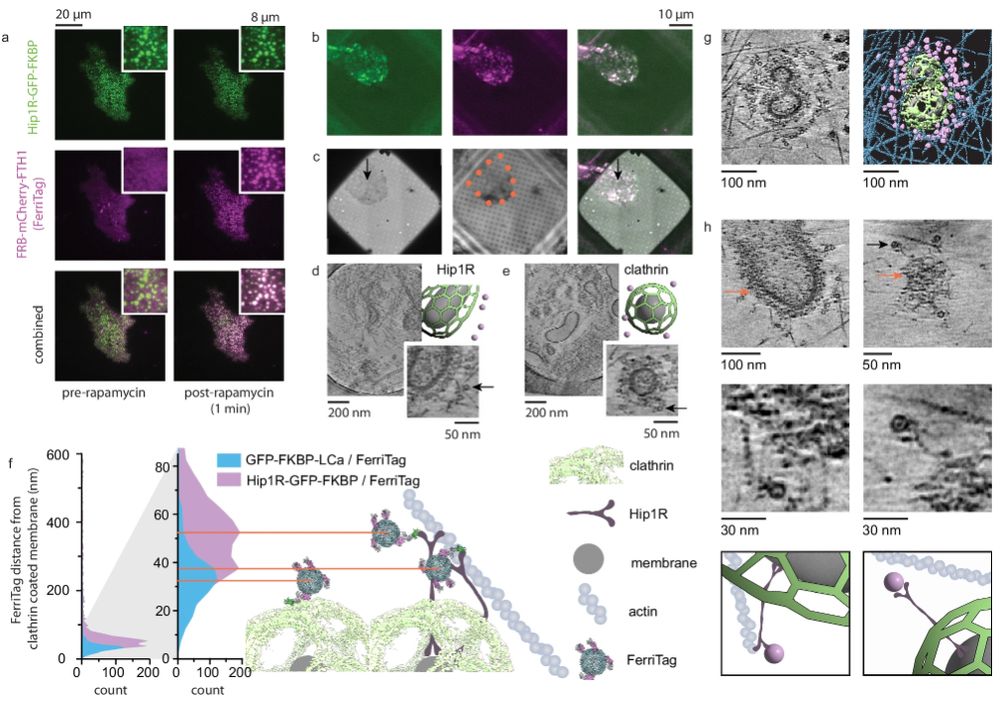
A realistic phantom dataset for benchmarking cryo-ET data annotation - Nature Methods
A standardized, realistic phantom dataset consisting of ground-truth annotations for six diverse molecular species is provided as a community resource for cryo-electron-tomography algorithm benchmarki...
doi.org
Reposted by Utz Ermel
Reposted by Utz Ermel
Reposted by Utz Ermel
Reposted by Utz Ermel
Utz Ermel
@uermel.bsky.social
· Mar 6
Reposted by Utz Ermel
Utz Ermel
@uermel.bsky.social
· Feb 28
Reposted by Utz Ermel
Reposted by Utz Ermel
Reposted by Utz Ermel
Reposted by Utz Ermel
cryoEM papers
@cryoempapers.bsky.social
· Feb 14
Reposted by Utz Ermel
Reposted by Utz Ermel
Reposted by Utz Ermel
Josh Moore
@joshmoore.bsky.social
· Jan 28
Reposted by Utz Ermel
Bridget Carragher
@bcarra.bsky.social
· Jan 23
Reposted by Utz Ermel
Reposted by Utz Ermel