Yasin El Abiead
@yelabiead.bsky.social
110 followers
120 following
14 posts
Interested in metabolomics, metabolism, and how to get from the former to the latter
Posts
Media
Videos
Starter Packs
Yasin El Abiead
@yelabiead.bsky.social
· Aug 27
Yasin El Abiead
@yelabiead.bsky.social
· Aug 27
Yasin El Abiead
@yelabiead.bsky.social
· Aug 27
Reposted by Yasin El Abiead
Reposted by Yasin El Abiead
BioMassSpec
@realbiomassspec.bsky.social
· Aug 25

Increasing the Scale of the Mass Spectrometry Query Language Compendium with Explainable AI
A significant bottleneck in metabolomics data interpretation is the effective use of domain knowledge to assign structural information based on fragmentation patterns. The mass spectrometry query lang...
pubs.acs.org
Yasin El Abiead
@yelabiead.bsky.social
· Aug 23

A Perspective on Unintentional Fragments and their Impact on the Dark Metabolome, Untargeted Profiling, Molecular Networking, Public Data, and Repository Scale Analysis.
In/post-source fragments (ISFs) arise during electrospray ionization or ion transfer in mass spectrometry when molecular bonds break, generating ions that can complicate data interpretation. Although ISFs have been recognized for decades, their contribution to untargeted metabolomics - particularly in the context of the so-called “dark matter” (unannotated MS or MS/MS spectra) and the “dark metabolome” (unannotated molecules) - remains unsettled. This ongoing debate reflects a central tension: while some caution against overinterpreting unidentified signals lacking biological evidence, others argue that dismissing them too quickly risks overlooking genuine molecular discoveries. These discussions also raise a deeper question: what exactly should be considered part of the metabolome? As metabolomics advances toward large-scale data mining and high-throughput computational analysis, resolving these conceptual and methodological ambiguities has become essential. In this perspective, we propose a refined definition of the “dark metabolome” and present a systematic overview of ISFs and related ion forms, including adducts and multimers. We examine their impact on metabolite annotation, experimental design, statistical analysis, computational workflows, and repository-scale data mining. Finally, we provide practical recommendations - including a set of dos and don’ts for researchers and reviewers - and discuss the broader implications of ISFs for how the field explores unknown molecular space. By embracing a more nuanced understanding of ISFs, metabolomics can achieve greater rigor, reduce misinterpretation, and unlock new opportunities for discovery.
doi.org
Reposted by Yasin El Abiead
Tri Kaloudis
@trikaloudis.bsky.social
· Aug 20

Reverse Spectral Search Reimagined: A Simple but Overlooked Solution for Chimeric Spectral Annotation
The exponential growth of untargeted metabolomics data, now reaching billions of mass spectra in public repositories, benefits from reannotation strategies for data reuse. While tandem mass spectromet...
doi.org
Reposted by Yasin El Abiead
unitsaq.bsky.social
@unitsaq.bsky.social
· Aug 14

Computationally unmasking each fatty acyl C=C position in complex lipids by routine LC-MS/MS lipidomics - Nature Communications
Physiologically relevant omega-positions of double bonds in fatty acyls in complex lipids can now only be identified with specialized instrumentation. Here, the authors present a computational approac...
www.nature.com
Reposted by Yasin El Abiead
Harsha Gouda
@harshagouda.bsky.social
· Aug 8

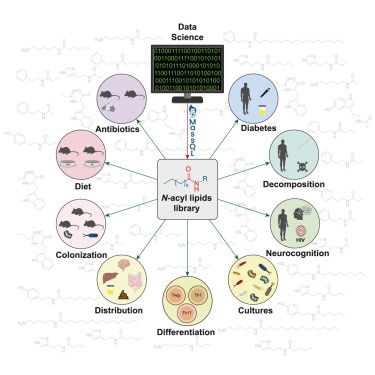
The mass spectrometry of microbiome-mediated metabolism of food: challenges and opportunities
With the exception of molecules acquired through the lungs, skin absorption, or part of a medication regime, nearly all molecules in our bodies origin…
www.sciencedirect.com
Reposted by Yasin El Abiead
Mingxun Wang
@mingxunwang.bsky.social
· Aug 2
Reposted by Yasin El Abiead
Reposted by Yasin El Abiead
Böckerlab
@boeckerlab.bsky.social
· Jul 28
Reposted by Yasin El Abiead
Daniel Petras
@daniel-petras.bsky.social
· Jul 27
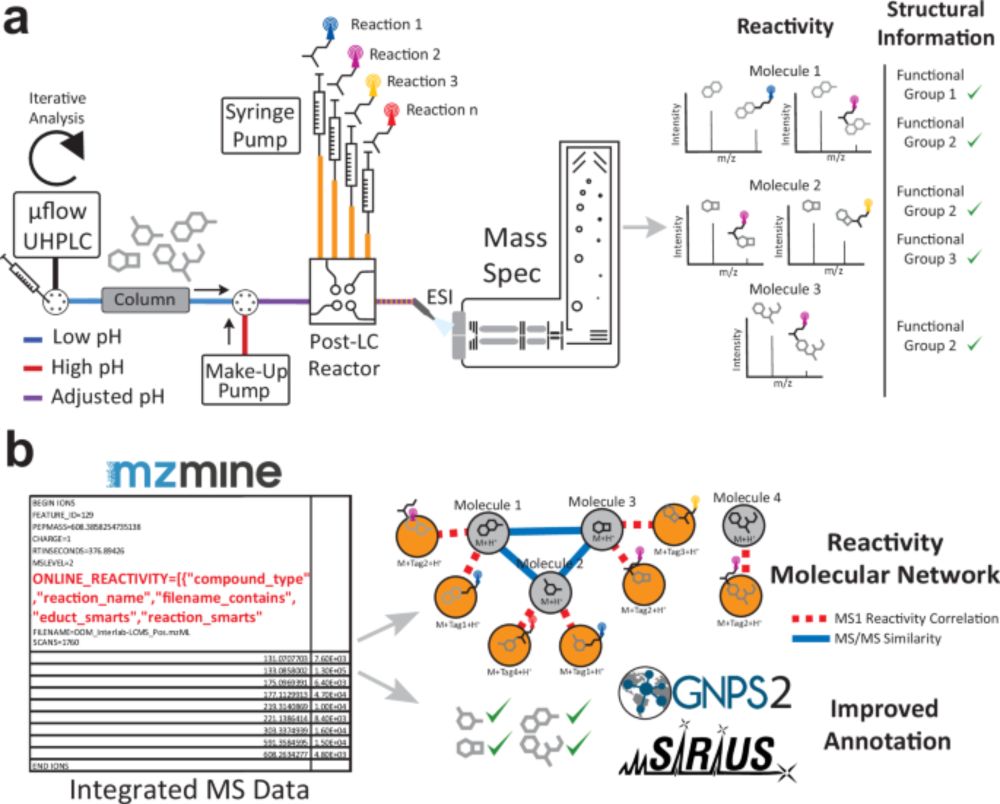
Enhancing tandem mass spectrometry-based metabolite annotation with online chemical labeling - Nature Communications
To improve annotation in non-targeted metabolomics studies, authors develop a Multiplexed Chemical Metabolomics (MCheM) platform, combining post-column derivatization with integrated data processing. ...
www.nature.com
Reposted by Yasin El Abiead
Steffen Neumann
@sneumann.bsky.social
· Jul 16
Reposted by Yasin El Abiead
BioMassSpec
@realbiomassspec.bsky.social
· Jul 13

An evaluation methodology for machine learning-based tandem mass spectra similarity prediction - BMC Bioinformatics
Background Untargeted tandem mass spectrometry serves as a scalable solution for the organization of small molecules. One of the most prevalent techniques for analyzing the acquired tandem mass spectr...
bmcbioinformatics.biomedcentral.com
Reposted by Yasin El Abiead
Reposted by Yasin El Abiead
Reposted by Yasin El Abiead
Yasin El Abiead
@yelabiead.bsky.social
· May 28
Reposted by Yasin El Abiead