Zach Marin
@zacsimile.bsky.social
540 followers
790 following
51 posts
hardware x software in microscopy | postdoc in Ries Lab @maxperutzlabs.bsky.social | http://zacsimile.github.io | 🇺🇸 in 🇦🇹 | he/him | views are graphical projections
Posts
Media
Videos
Starter Packs
Reposted by Zach Marin
Reposted by Zach Marin
Reposted by Zach Marin
Reposted by Zach Marin
Reposted by Zach Marin
Reposted by Zach Marin
Yiming Li
@yimingli.bsky.social
· Aug 6

Scalable and lightweight deep learning for efficient high accuracy single-molecule localization microscopy
Nature Communications - This study presents LiteLoc, a lightweight and scalable AI model for efficient and accurate single molecule localization microscopy data analysis, bringing real-time...
rdcu.be
Reposted by Zach Marin
Reposted by Zach Marin
Reposted by Zach Marin
Reposted by Zach Marin
Zach Marin
@zacsimile.bsky.social
· Apr 24

Sidelobe-free deterministic 3D nanoscopy with λ/33 axial resolution - Light: Science & Applications
A purely physical, single objective-lens strategy (UNEx-4Pi) for achieving sidelobe-free λ/33 axial resolution by fusion of ultrahighly nonlinear excitation of photon avalanches and mirror-assisted bi...
www.nature.com
Reposted by Zach Marin
Rita Strack
@ritastrack.bsky.social
· Apr 10
Zach Marin
@zacsimile.bsky.social
· Apr 10
Reposted by Zach Marin
Christophe 🔬 L
@christlet.bsky.social
· Apr 9

Advancing Super-Resolution Microscopy: Recent Innovations in Commercial Instruments
Abstract. Super-resolution microscopy techniques have accelerated scientific progress, enabling researchers to explore cellular structures and dynamics wit
academic.oup.com
Reposted by Zach Marin










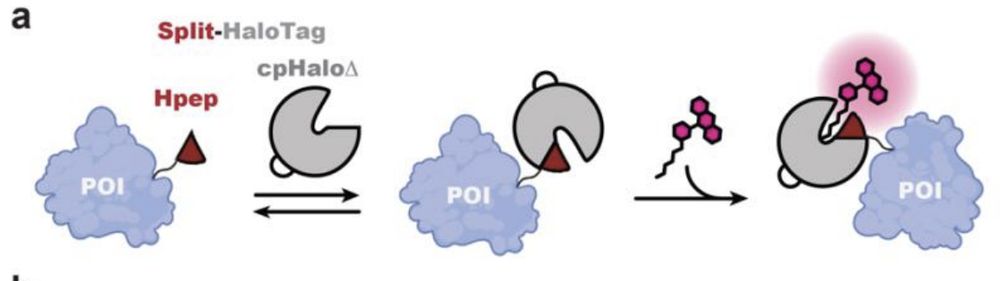

![Live-cell confocal imaging of histone H2B type-2E-Hpep11 CRISPR KI cell lines after 2-hours labeling with the specified CA- ligand [100 nM]. Images were taken with optimal image acquisition parameter for each dye. Scale bar: 50 μm.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:uf4m7zlmcjlg2biwn4s2k32y/bafkreibozn5fz6sgeksqrz3vmy6uiydtnvwmb5my7r3xecko2erokryn3a@jpeg)
![(a) Confocal laser scanning microscopy (CLSM) and STED images of mitochondria in U2OS cells coexpressing cpHaloΔ3 and TOM20-Hpep, either overexpressed or endogenously tagged. Scale bar: 1 μm. Pixel intensities scaled according to reference bar. (b) Representative CLSM, STED images of the CRISPR/Cas9 KI cells expressing TOM20 tagged with intact HaloTag (upper) or Hpep11 (bottom). (c) Intensity profiles along mitochondrial tubules (red and blue lines in b). Scale bar: 2 μm. (d) Representative CLSM and STED images of endogenous Hpep11-tagged clathrin with cpHaloΔ3 coexpression. Scale bar: 10 μm (overview) and 2 μm (magnification). (e) Representative CLSM, STED
images of endogenously tagged tubulin beta 4B with Hpep11. Scale bar: 10 μm (overview) and 2 μm (magnification). (f) Intensity profiles along tubulin filaments (red and blue lines in e) Means ± s.d. of the filament diameters were calculated as full width at half maximum (FWHM) from n=20 microtubule filaments, ≥ 2 images. A slight increase in cytosolic signal was noted in cells tagged with split-HaloTagat TUBB4B, compared to cells tagged with the full-length HaloTag, which may result from the presence of unbound but labeled cpHaloΔ3. All images were acquired after labeling with CA-SiR [100 nM] for one hour.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:uf4m7zlmcjlg2biwn4s2k32y/bafkreielty6fex37xy4caechbw5ebo3eypyiyvxxhkigeazd5gmy6tvlay@jpeg)