
MS, Geological Sciences
BS, Neuroscience
Founder of MAB:
midauthorbio.com
GitHub:
https://github.com/Arkadiy-Garber
https://github.com/Middle-Author-Bioinformatics
Google Scholar:
https://scholar.google.com/citations?user=SGPloYgAAA
A bile acid-bound structure of toxin TcdB revealed the mechanism of inhibition and guided the design of a synthetic bile acid that alleviated Clostridioides difficile infection in mice.
#MicroSky 🦠
www.nature.com/articles/s41...

A bile acid-bound structure of toxin TcdB revealed the mechanism of inhibition and guided the design of a synthetic bile acid that alleviated Clostridioides difficile infection in mice.
#MicroSky 🦠
www.nature.com/articles/s41...

doi.org/10.1093/bioa...
The GlobDB is the largest species dereplicated genome database currently available, containing 306,260 species representatives.
More information on globdb.org 1/5
🖥️🧬🦠

doi.org/10.1093/bioa...
The GlobDB is the largest species dereplicated genome database currently available, containing 306,260 species representatives.
More information on globdb.org 1/5
🖥️🧬🦠

nature.com/articles/s41...
Thank you, Jan, Danny (@dannyjamesward.bsky.social), Joana (@joanampereira.bsky.social), as well as reviewers and editors @natrevmicro.nature.com!

nature.com/articles/s41...
Thank you, Jan, Danny (@dannyjamesward.bsky.social), Joana (@joanampereira.bsky.social), as well as reviewers and editors @natrevmicro.nature.com!
Review on the T6SS structure and effectors.
journals.asm.org/doi/10.1128/...

Review on the T6SS structure and effectors.
journals.asm.org/doi/10.1128/...
Deadline: Dec. 2, 2 p.m. ET.
Share your science → asm.social/2Gv

Deadline: Dec. 2, 2 p.m. ET.
Share your science → asm.social/2Gv

We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
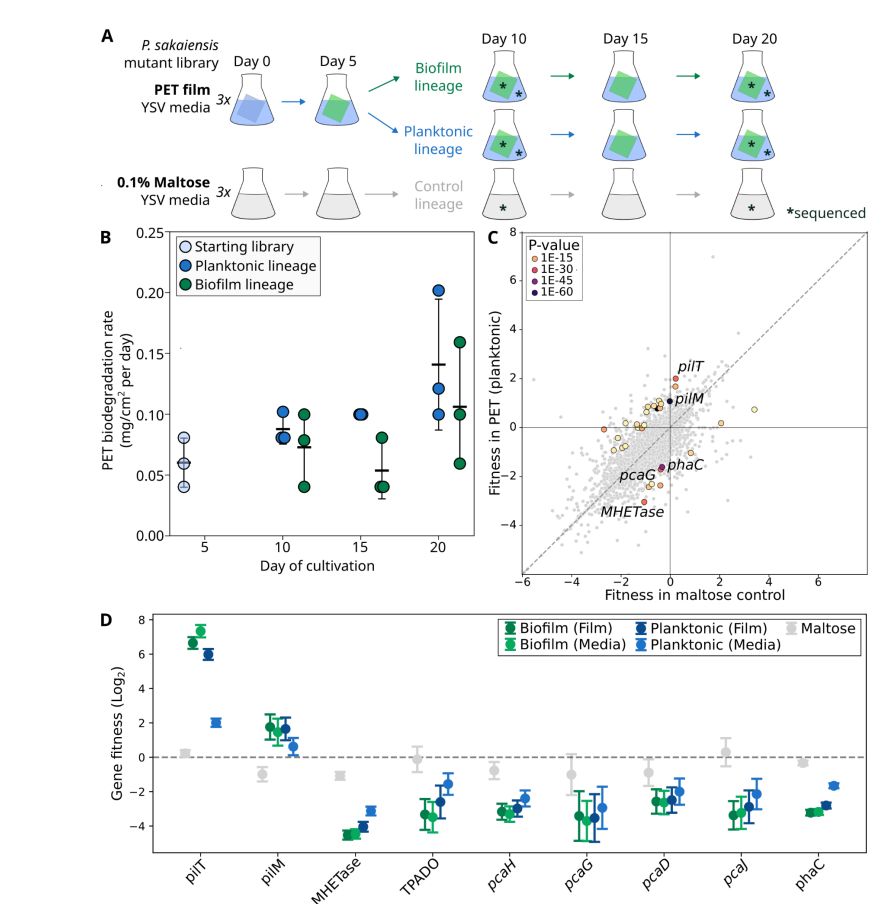
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
Primary Plastid in the Making”. This is cool as hell 😎
www.biorxiv.org/content/bior...
Primary Plastid in the Making”. This is cool as hell 😎
www.biorxiv.org/content/bior...

Review on the T6SS structure and effectors.
journals.asm.org/doi/10.1128/...

Review on the T6SS structure and effectors.
journals.asm.org/doi/10.1128/...
🦠🧪
www.nature.com/articles/d41...

rdcu.be/eLtCH

rdcu.be/eLtCH



