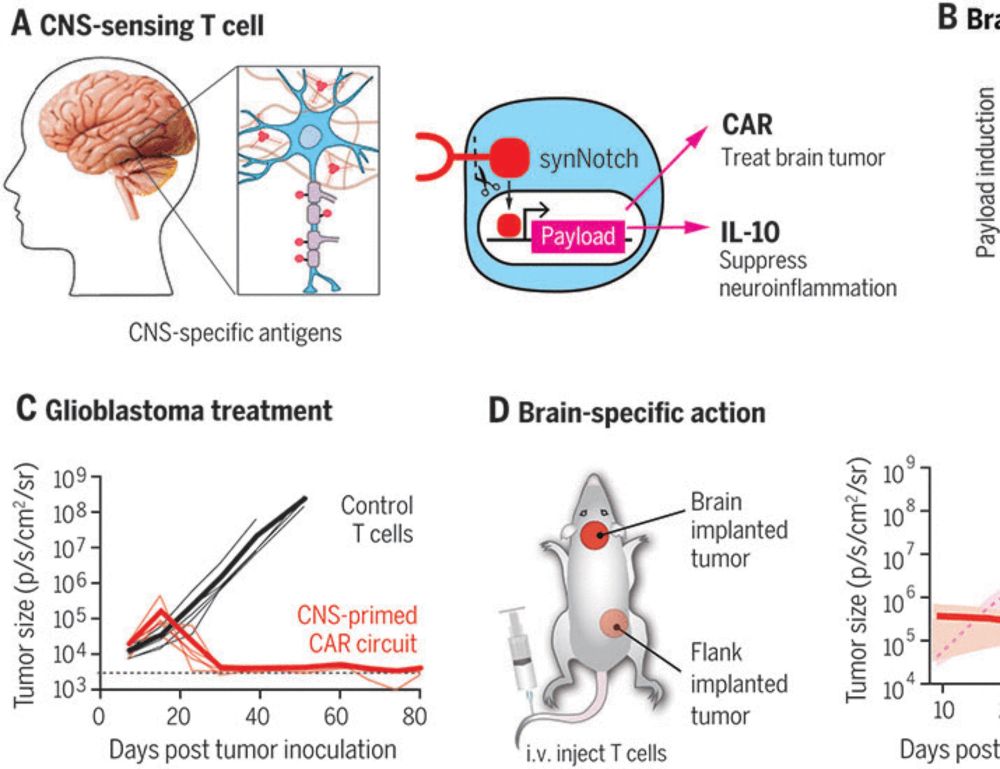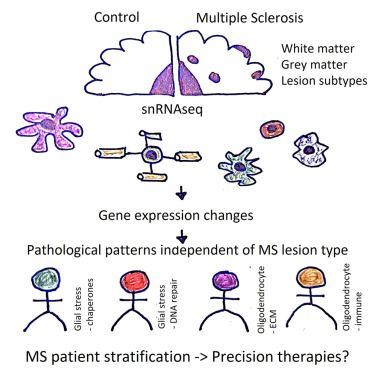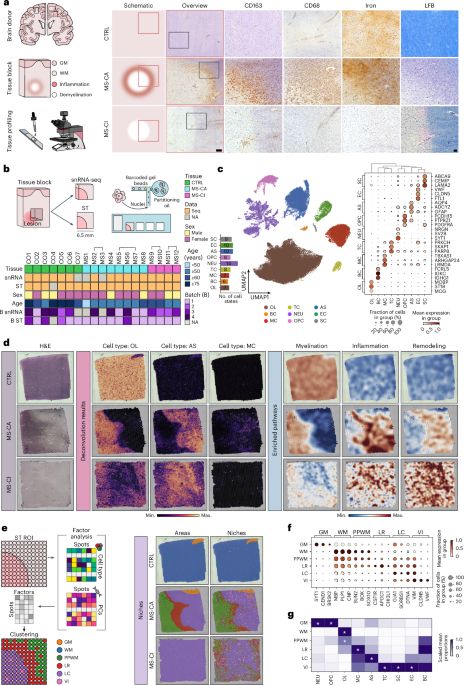
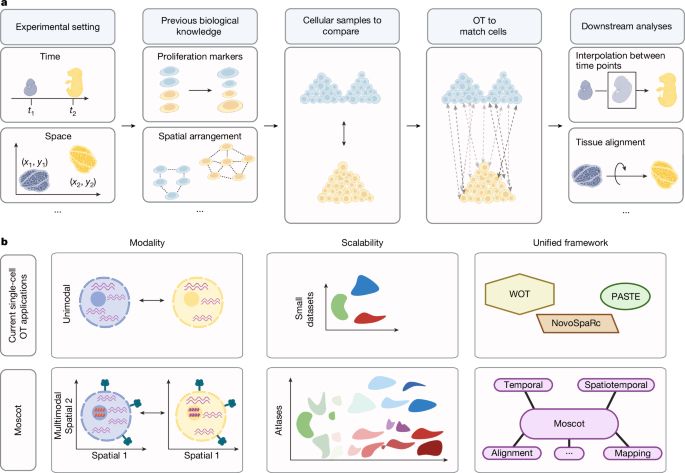
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
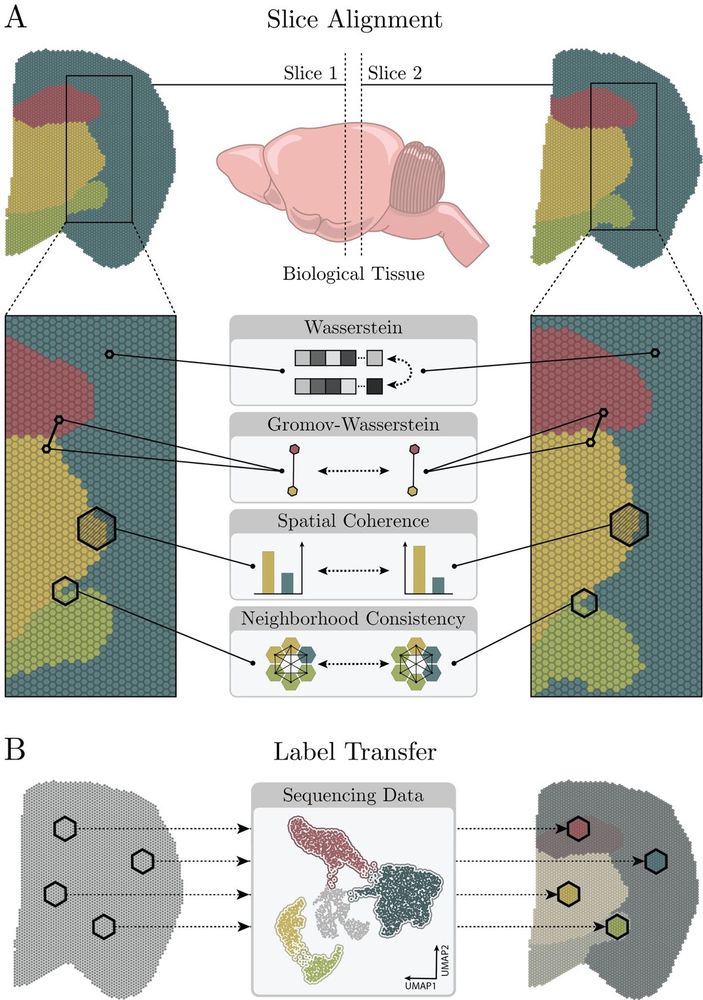
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
@sciimmunology.bsky.social
www.science.org/doi/10.1126/...
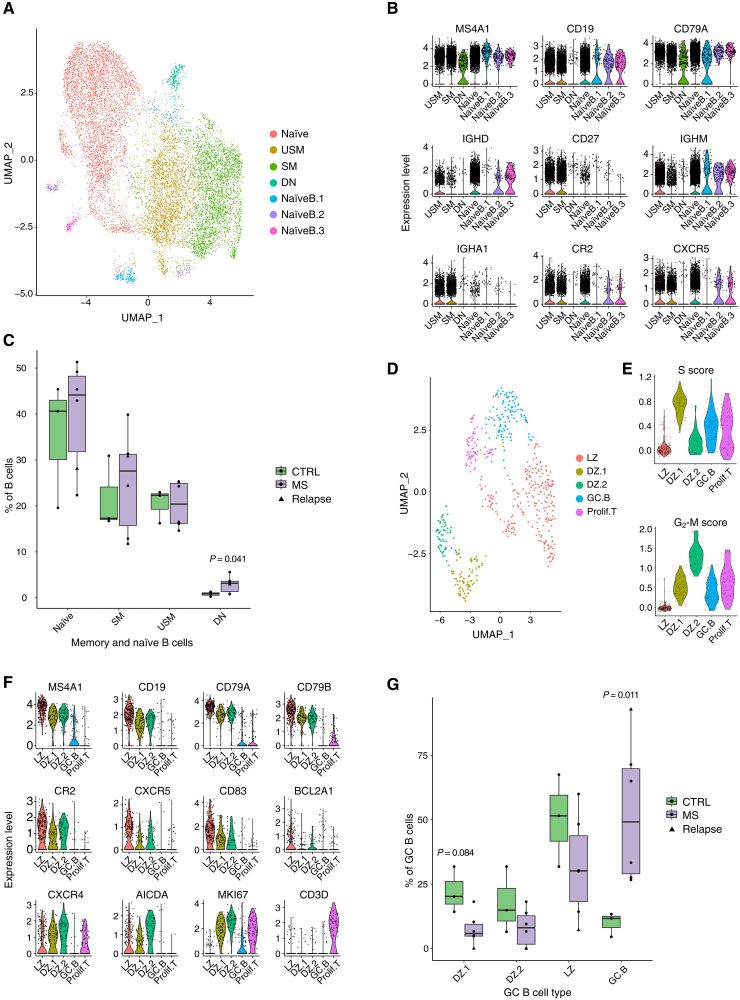
@sciimmunology.bsky.social
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
TL;DR: BCRs ARE ALL YOU NEED!
(Well actually .... keep reading) 1/

www.nature.com/articles/s41...
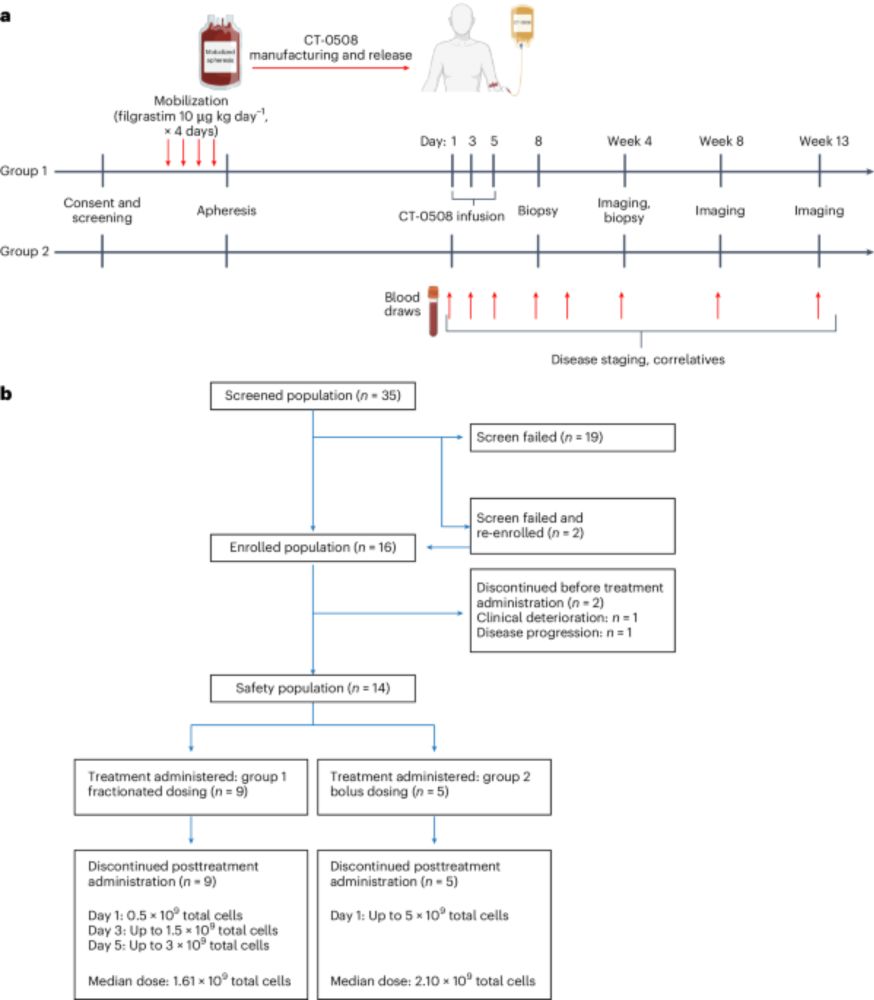
www.nature.com/articles/s41...
Learn more about the Solve-RD project and how it aims to collect and harmonise #raredisease data across multiple European partners.
🧬🖥️
www.ebi.ac.uk/about/news/a...
Learn more about the Solve-RD project and how it aims to collect and harmonise #raredisease data across multiple European partners.
🧬🖥️
www.ebi.ac.uk/about/news/a...
www.nature.com/articles/s41...
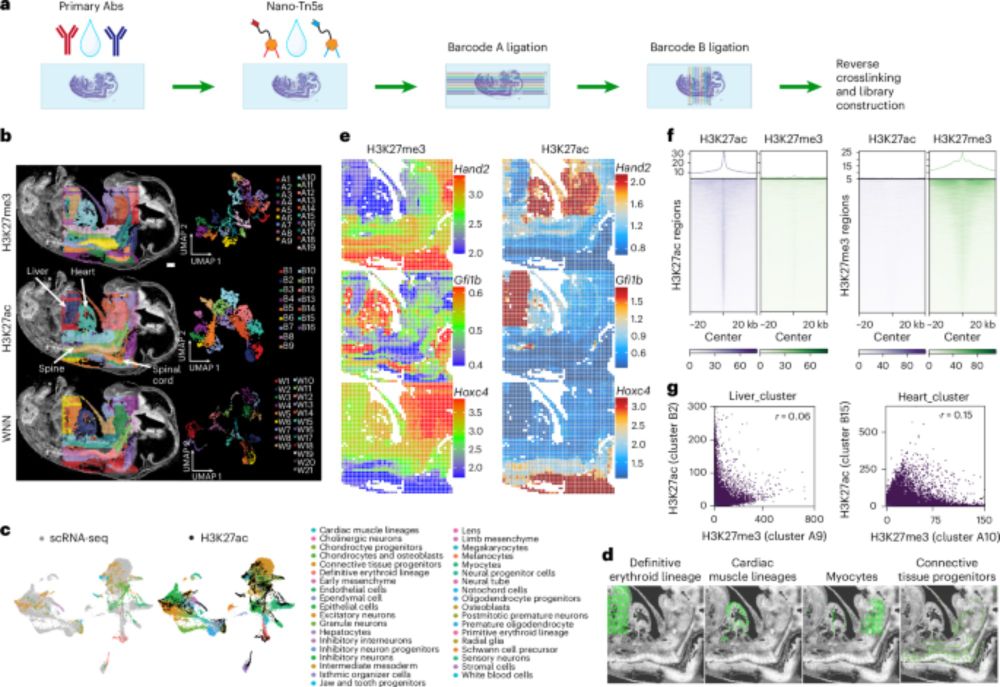
www.nature.com/articles/s41...
@naturecomms.bsky.social
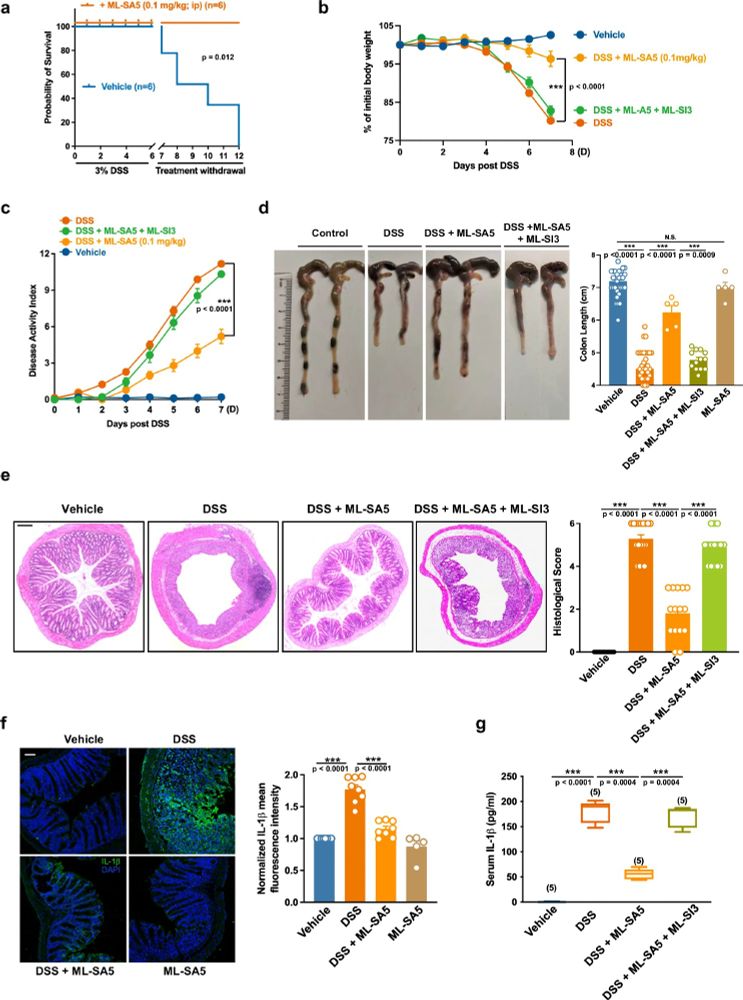
www.nature.com/articles/s41...
@naturecomms.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.nature.com/articles/s41...
For more on the remarkable surge of LLLMs see erictopol.substack.com/p/learning-t...
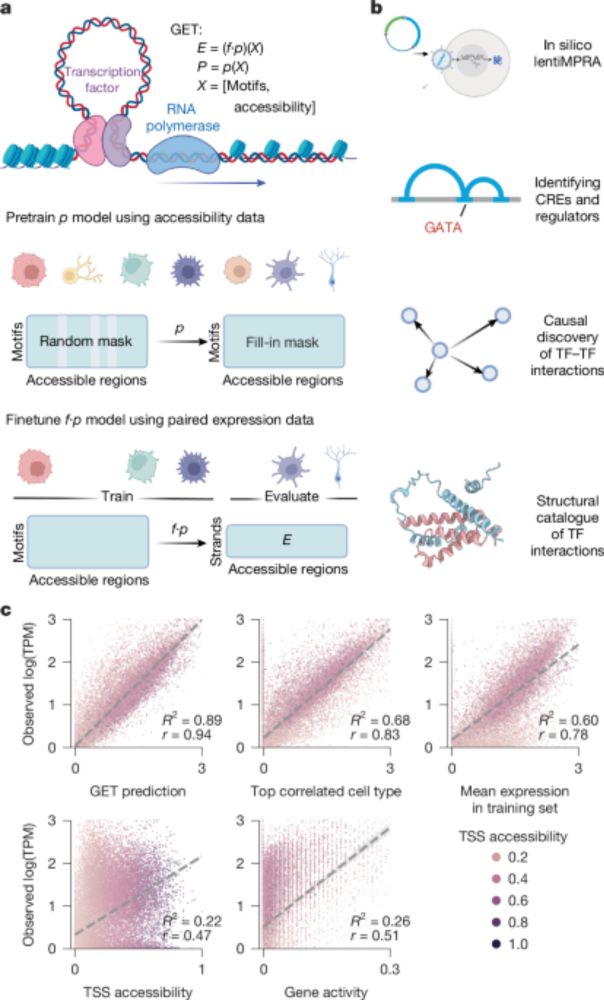
For more on the remarkable surge of LLLMs see erictopol.substack.com/p/learning-t...
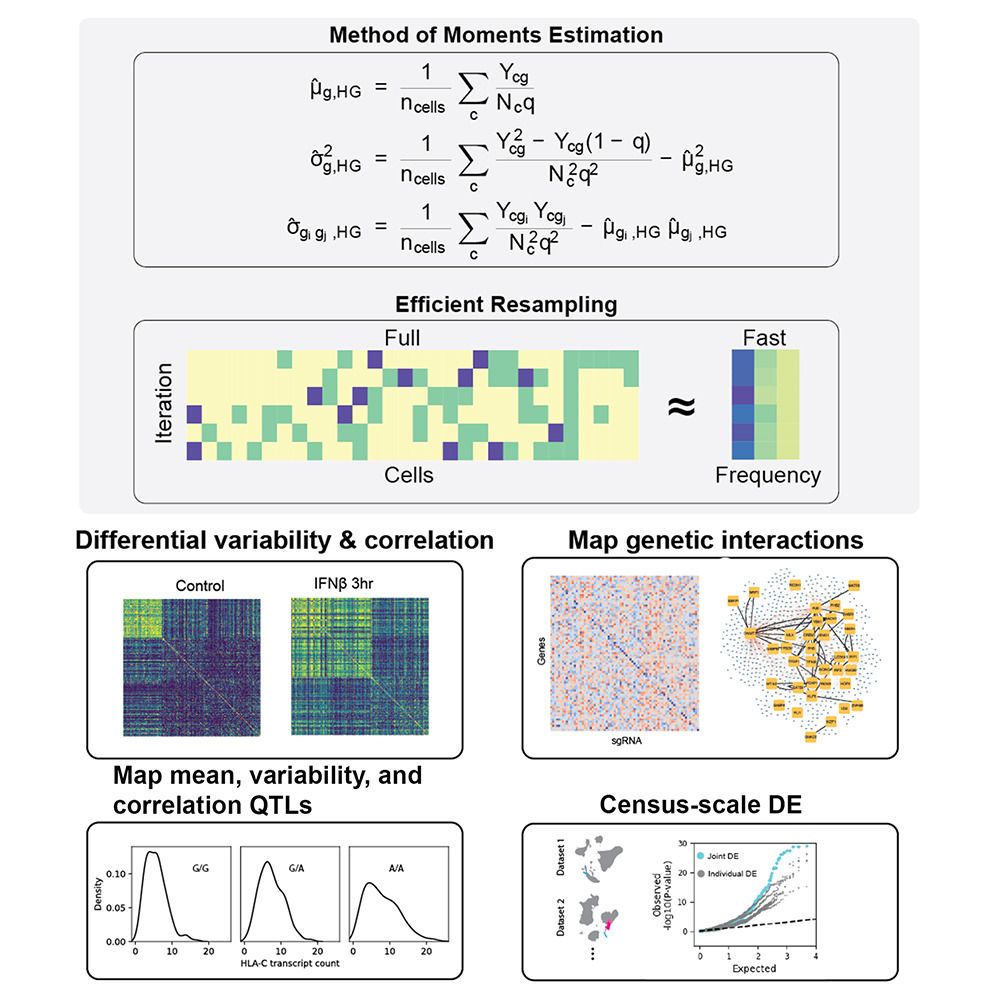
doi.org/10.1038/s415...
doi.org/10.1038/s415...
www.nature.com/articles/s41...
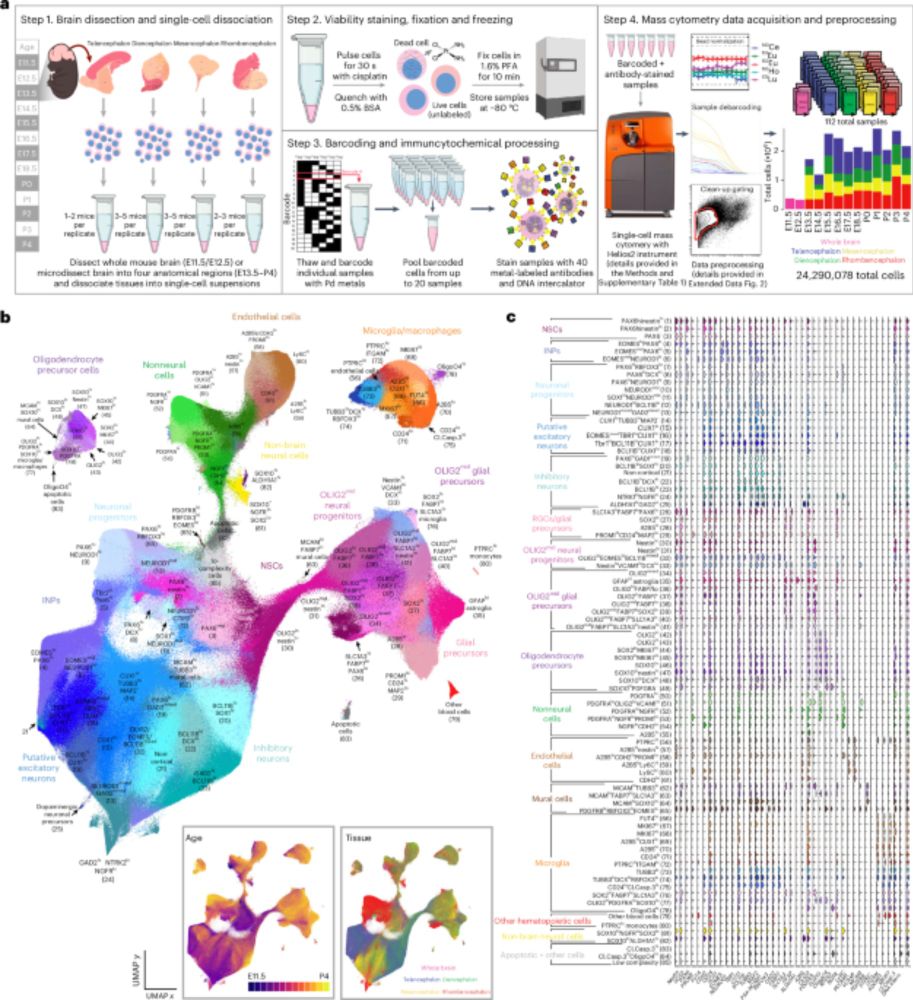
www.nature.com/articles/s41...
www.nature.com/articles/s41...
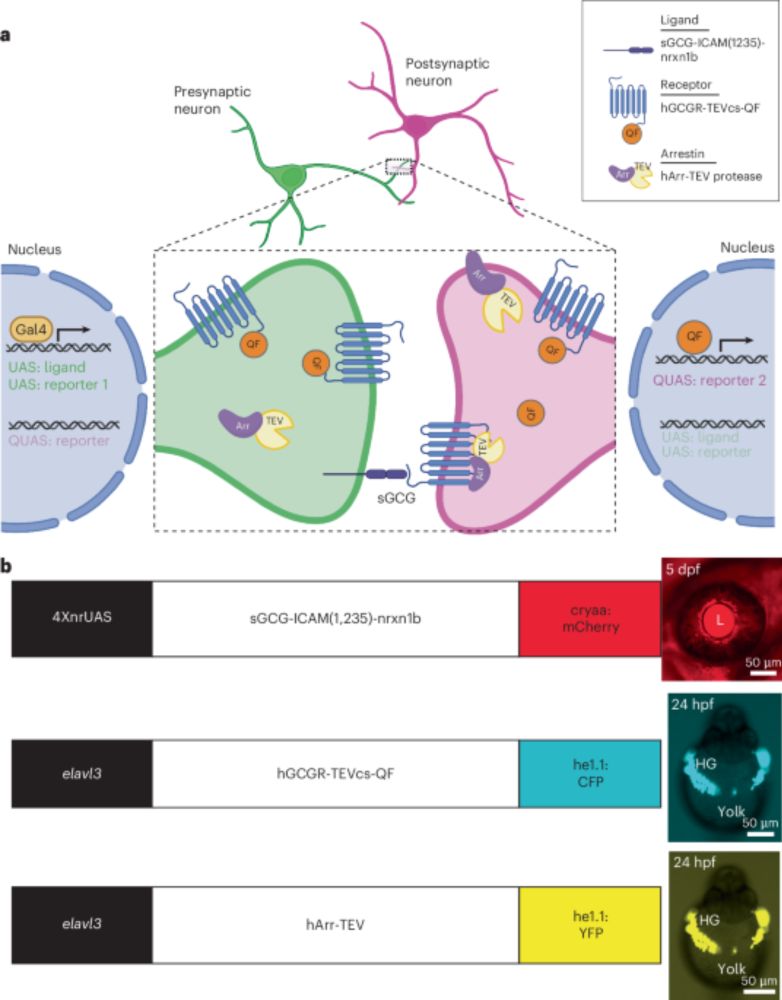
www.nature.com/articles/s41...
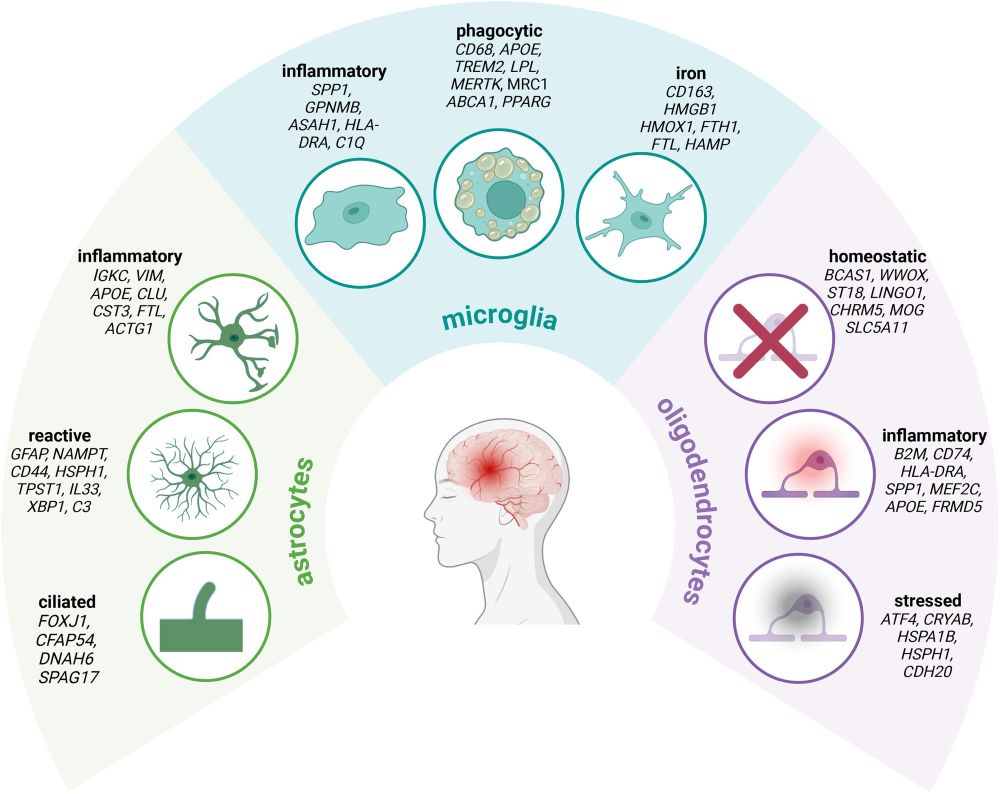
www.biorxiv.org/content/10.1...
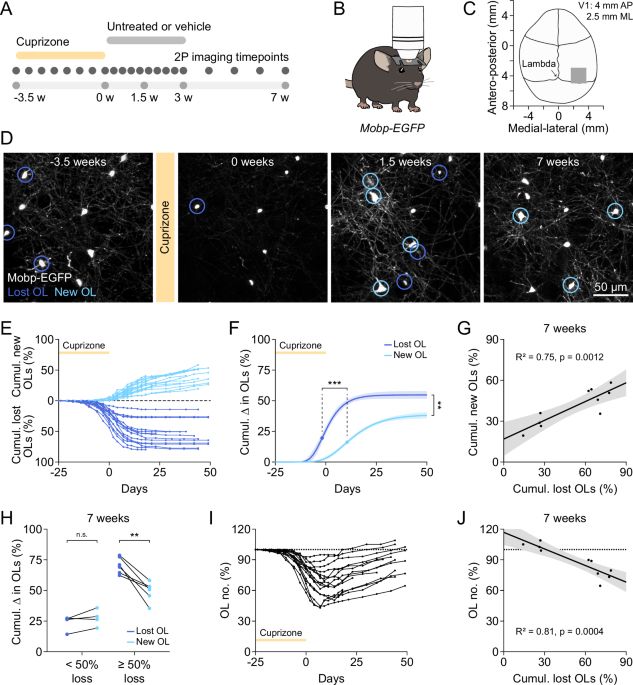
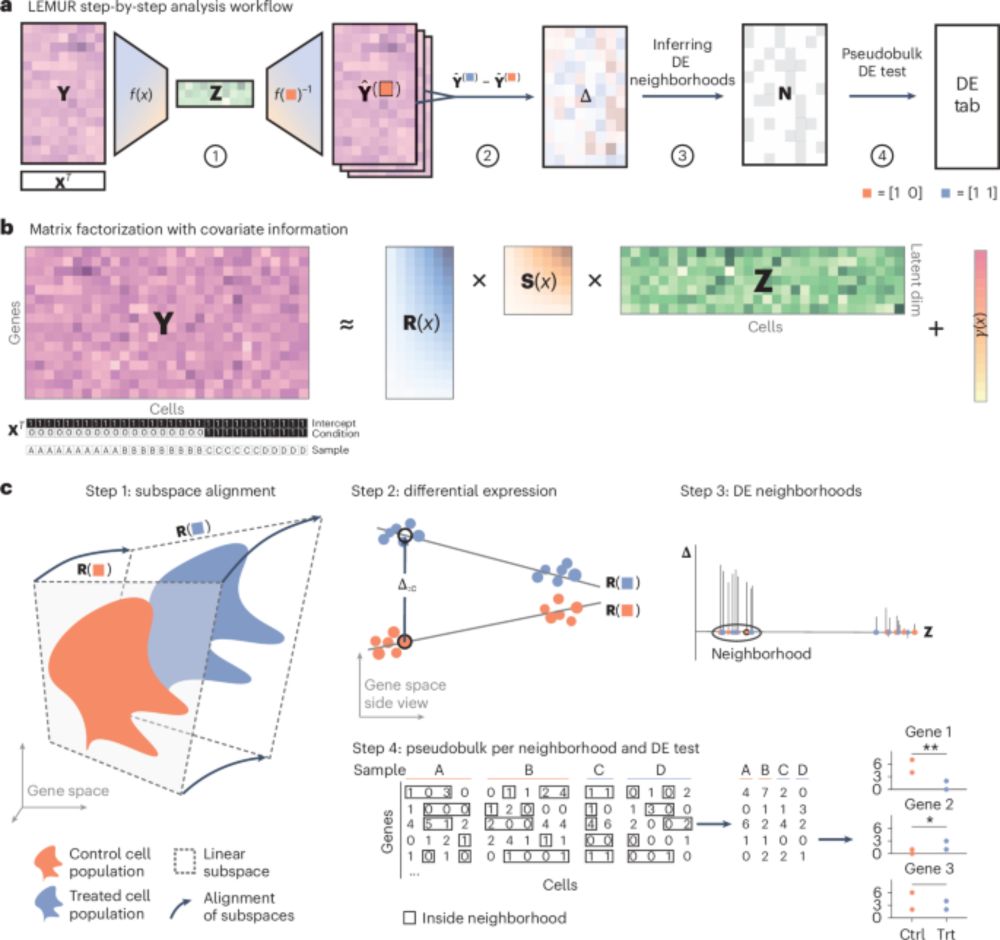
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
@annawilliamslab.bsky.social
@willmacnair.bsky.social
@deckardw.bsky.social
@karolinskainst.bsky.social
@edinburghuni.bsky.social
@cp-neuron.bsky.social
and many others....

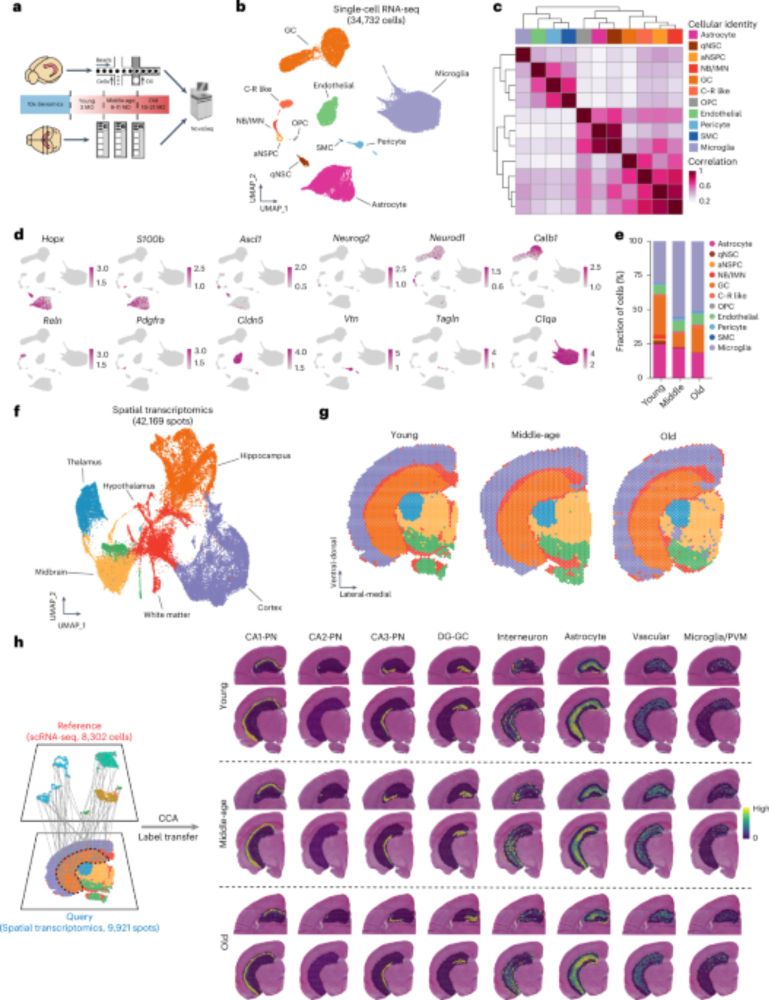
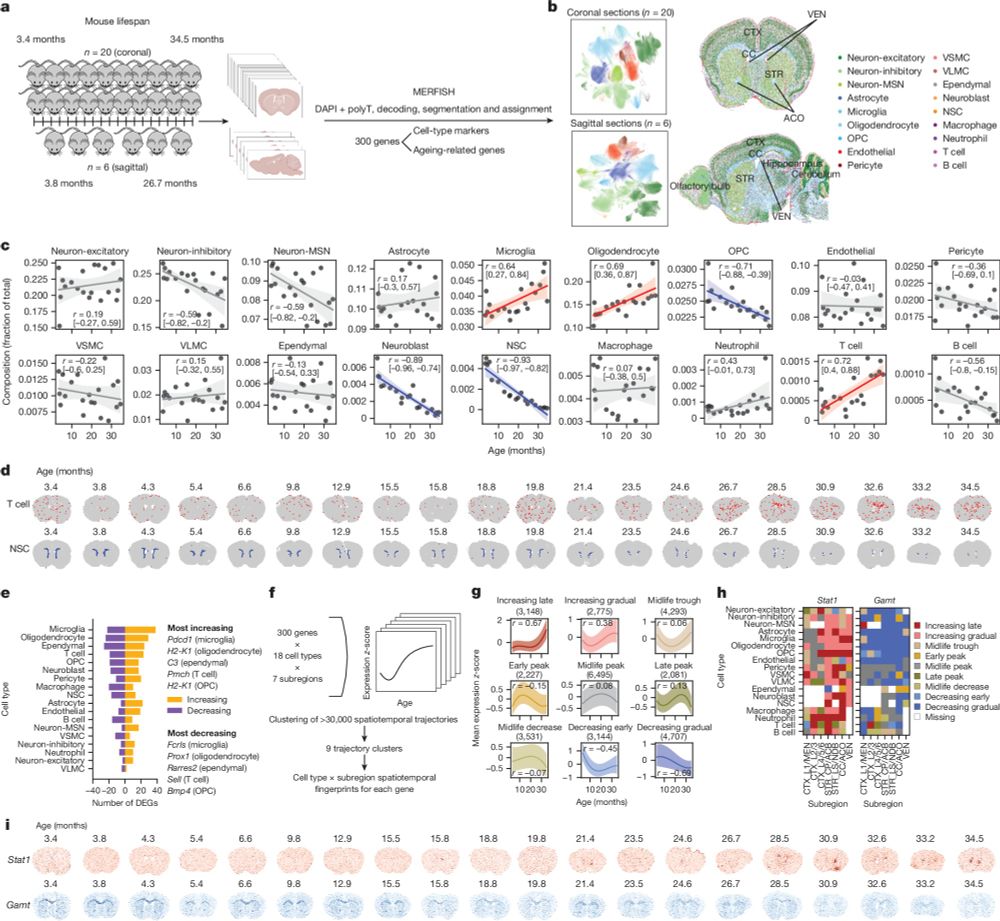
Spatial transcriptomics of brain aging and 'spatial aging clocks' identify cells that have pro-aging or pro-rejuvenating effects on their neighbors!
Huge CONGRATS to Eric Sun and all authors! Fantastic collaboration with @jameszou.bsky.social!
rdcu.be/d33uQ 🧵
👉🏽with Gaurav Chopra, Caitlin Randolph, and @katherinewalker.bsky.social
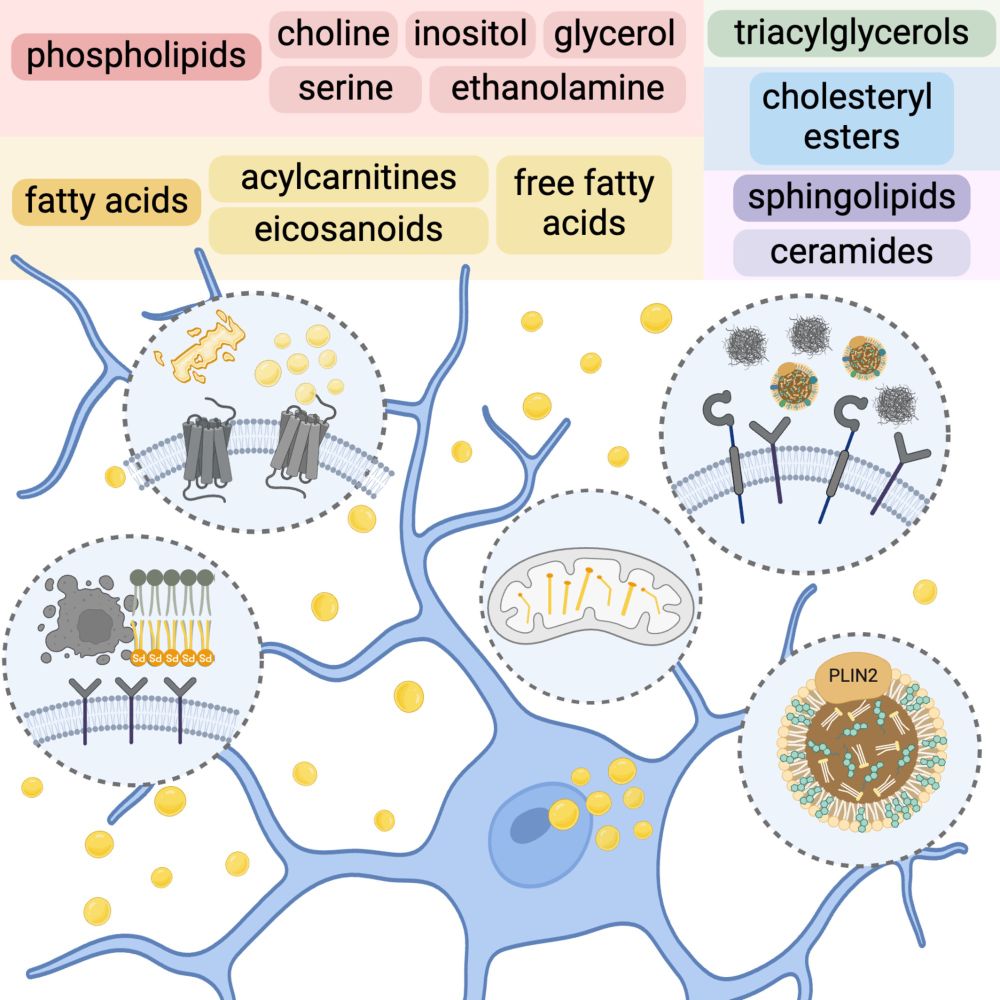
👉🏽with Gaurav Chopra, Caitlin Randolph, and @katherinewalker.bsky.social