Geoff Faulkner
@faulknerlab.bsky.social
450 followers
120 following
44 posts
Professor, University of Queensland. Retrotransposons and genomics.
Views my own. Scholar publications: http://tinyurl.com/3k7r3xvj
Posts
Media
Videos
Starter Packs
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Johan Jakobsson
@jakobssonlab.bsky.social
· Aug 23
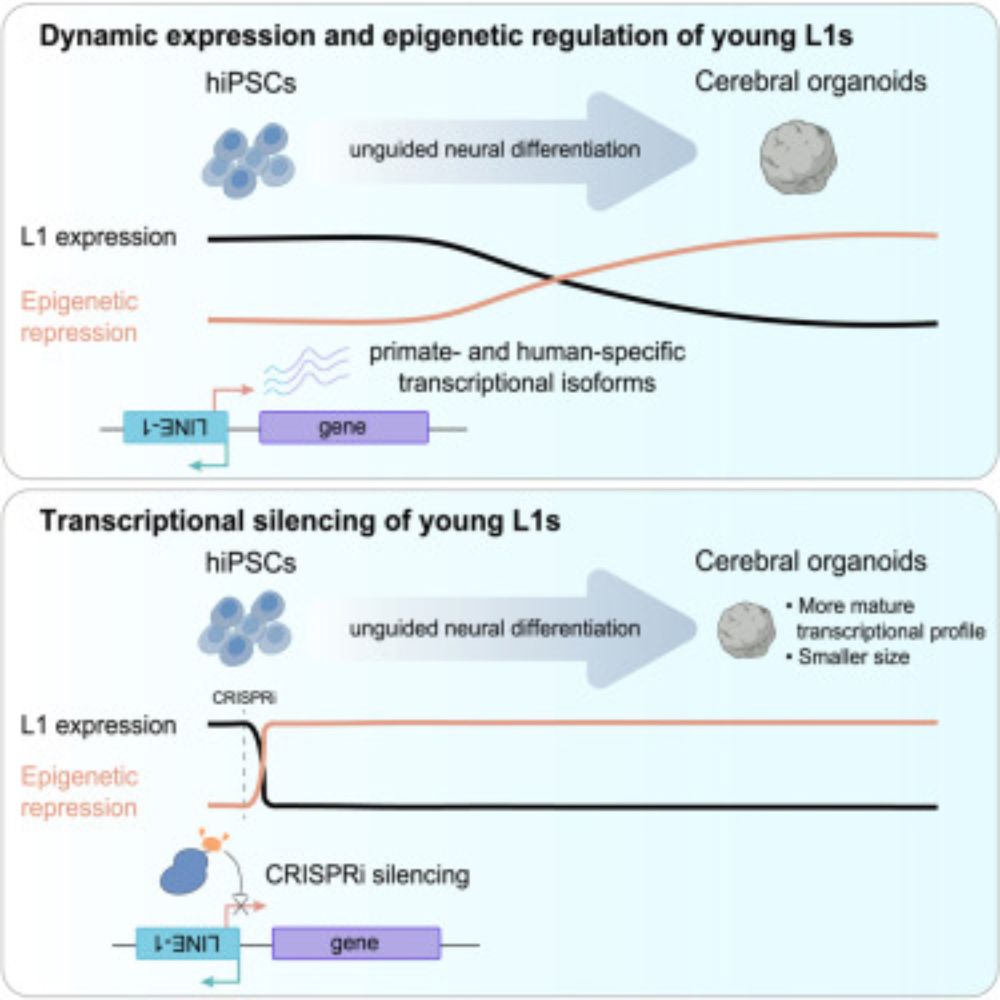
LINE-1 retrotransposons mediate cis-acting transcriptional control in human pluripotent stem cells and regulate early brain development
Adami et al. demonstrate that evolutionarily young L1s are expressed in human pluripotent
stem cells and are dynamically regulated throughout neural differentiation. The study
examines the role of L1s...
www.cell.com
Reposted by Geoff Faulkner
Miguel Branco
@brancolab.bsky.social
· Aug 22

ADAR1 as a Placental Innate Immune Rheostat Sustaining the Homeostatic Balance of Intrinsic Interferon Response at the Maternal‐Fetal Interface
This study reveals that ADAR1, an RNA-editing enzyme, fine-tunes immune responses in the placenta by preventing the accumulation of immunogenic double-stranded RNAs (dsRNAs) from interferon-stimulate...
advanced.onlinelibrary.wiley.com
Reposted by Geoff Faulkner
Marco Trizzino
@marcotrizzino.bsky.social
· Aug 20
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Maxim Greenberg
@maxvcg.bsky.social
· Aug 9

UHRF2 mediates resistance to DNA methylation reprogramming in primordial germ cells - Nature Communications
DNA methylation in mouse primordial germ cells (PGCs) is restricted to transposable elements, but how this unique DNA methylome is established is poorly understood. Here, the authors identify UHRF2 as...
www.nature.com
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Rebecca Berrens
@rberrens.bsky.social
· Jul 16
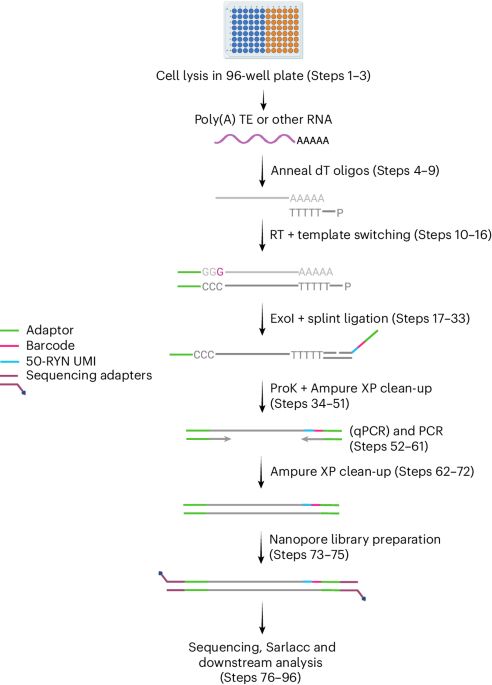
Long-read RNA sequencing of transposable elements from single cells using CELLO-seq - Nature Protocols
Single-cell long-read RNA sequencing enables the high-fidelity mapping of single-cell expression data from highly sequence-similar transposable elements to unique genomic loci by correcting errors fro...
doi.org
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Tugce Aktas
@aktast.bsky.social
· Jun 30
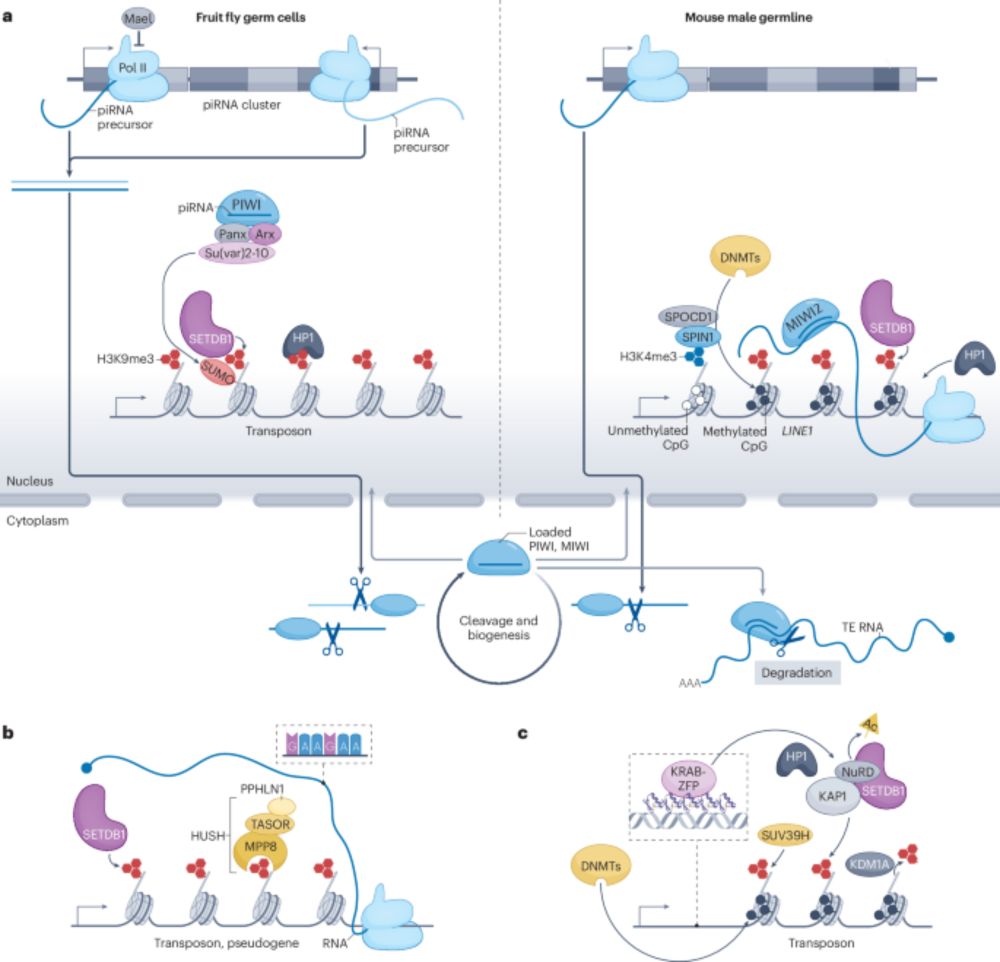
Transcriptional and post-transcriptional regulation of transposable elements and their roles in development and disease - Nature Reviews Molecular Cell Biology
Transposable elements (TEs) comprise nearly half of the human genome. This Review discusses transcriptional and post-transcriptional mechanisms that repress TE activity, how TEs escape this suppressio...
www.nature.com
Reposted by Geoff Faulkner
Reposted by Geoff Faulkner
Kasia Siudeja
@ksiudeja.bsky.social
· Jun 18
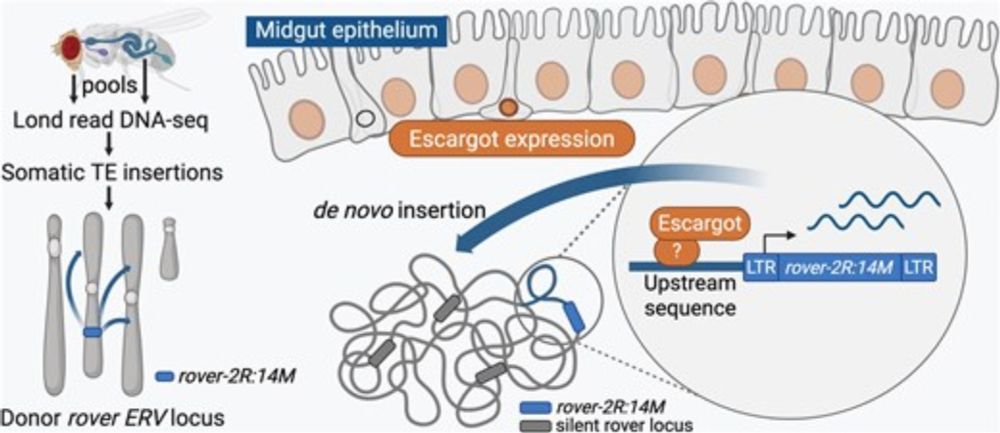
An endogenous retroviral element co-opts an upstream regulatory sequence to achieve somatic expression and mobility
Abstract. Retrotransposons, multi-copy sequences that propagate via copy-and-paste mechanisms, occupy large portions of eukaryotic genomes. A great majorit
doi.org